
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
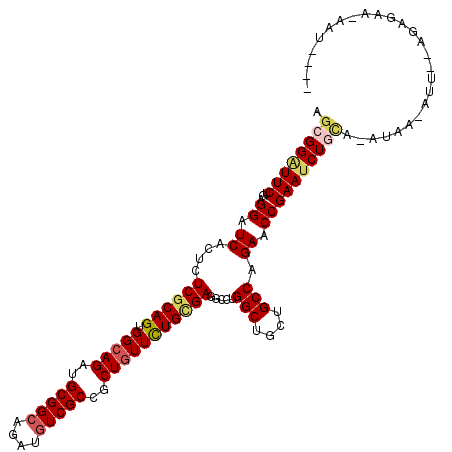
| Location | 9,364,658 – 9,364,771 |
| Length | 113 |
| Max. P | 0.803617 |

| Location | 9,364,658 – 9,364,771 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.62 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9364658 113 + 23771897 AGCGGAUUCUGCGGAUCACUCUCGCAGUGGCAGAUGCGGCAGAUGUCGCCGCUGUUCUGCGAUGCCUGGCUGCUGCCCGAACCGAAUCUGGA-AGGG-AUU--AGAGCAGGAUUAGA ....((((((((.((((....((((((.(((((..(((((....)))))..)))))))))))..((((((....)))....(((....))).-))))-)))--...))))))))... ( -44.50) >DroVir_CAF1 3025 114 + 1 AGCGGAUUCUGUGGAUCGCUCUCGCAGUGGCAGAUGCGGCAAAUGUCGCCGCUAUUCUGUGACGCCUGGCUGCUGCCAGAGCCGAAUCUGCGAAUAA-AUU--UGGACACGAUUAAA .((((((((((((((....)).))))((.(((((.(((((.......)))))...))))).))(((((((....))))).)).))))))))......-...--.............. ( -46.10) >DroPse_CAF1 8552 108 + 1 AGAGGGUUCGAAGGAUCACUCUCGCAGUGGCAGAUGCGGCAGAUGUCGCCGCUGUUUUGCGAGGCCUGGCUGCUGCCAGAACCGAAUCUGCA-AUAG-AGC--ACAGAG-AAC---- ...(((((((.........((((((((.(((((..(((((....)))))..))))))))))))).(((((....)))))...)))))))((.-....-.))--......-...---- ( -41.40) >DroMoj_CAF1 3737 114 + 1 AGCGGAUUCUGUGGAUCGCUCUCGCAGUGGCAGAUGCGGCAUAUAUCGCCGCUGUUCUGUGACGCUUGGCUGCUGCCCGAGCCGAAUCUGCA-AUAA-AUU-GUGAGCUCCAUUAUA .((((((((((((((....)).))))((.(((((.(((((.......)))))...))))).))(((((((....).)))))).)))))))).-....-..(-((((......))))) ( -43.50) >DroAna_CAF1 2603 109 + 1 AGCGGAUUCUGGGGAUCGCUCUCGCAGUGGCAGAUGCGGCAGAUGUCGCCGCUGUUCUGGGAGGCCUGGCUGCUGCCCGAACCGAACCUGUA-AGGAUAUUUCAGAGAA---U---- ......((((((((.((((.(((.(((.(((((..(((((....)))))..)))))))).)))))..(((....))).)).))...((....-.)).....))))))..---.---- ( -38.10) >DroPer_CAF1 13509 108 + 1 AGAGGGUUCGAAGGAUCACUCUCGCAGUGGCAGAUGCGGCAGAUGUCGCCGCUGUUUUGCGAGGCCUGGCUGCUGCCAGAACCGAAUCUGCA-AUAG-AGC--ACAGAG-AAU---- ...(((((((.........((((((((.(((((..(((((....)))))..))))))))))))).(((((....)))))...)))))))((.-....-.))--......-...---- ( -41.40) >consensus AGCGGAUUCUGAGGAUCACUCUCGCAGUGGCAGAUGCGGCAGAUGUCGCCGCUGUUCUGCGAGGCCUGGCUGCUGCCAGAACCGAAUCUGCA_AUAA_AUU__AGAGAA_AAU____ .((((((((...((.((....((((((.(((((..(((((....)))))..))))))))))).....(((....))).)).)))))))))).......................... (-33.12 = -33.62 + 0.50)

| Location | 9,364,658 – 9,364,771 |
|---|---|
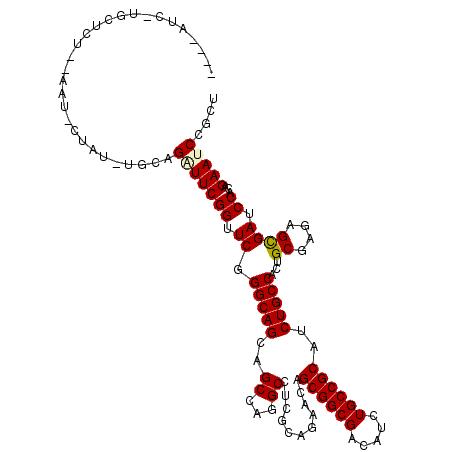
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9364658 113 - 23771897 UCUAAUCCUGCUCU--AAU-CCCU-UCCAGAUUCGGUUCGGGCAGCAGCCAGGCAUCGCAGAACAGCGGCGACAUCUGCCGCAUCUGCCACUGCGAGAGUGAUCCGCAGAAUCCGCU .......(((((..--...-(((.-.((......))...))).)))))...(((...(((((...((((((.....)))))).)))))..(((((.((....))))))).....))) ( -39.80) >DroVir_CAF1 3025 114 - 1 UUUAAUCGUGUCCA--AAU-UUAUUCGCAGAUUCGGCUCUGGCAGCAGCCAGGCGUCACAGAAUAGCGGCGACAUUUGCCGCAUCUGCCACUGCGAGAGCGAUCCACAGAAUCCGCU ..............--...-...(((((((....(((((((((....)))))).))).((((...(((((((...))))))).))))...)))))))((((............)))) ( -38.90) >DroPse_CAF1 8552 108 - 1 ----GUU-CUCUGU--GCU-CUAU-UGCAGAUUCGGUUCUGGCAGCAGCCAGGCCUCGCAAAACAGCGGCGACAUCUGCCGCAUCUGCCACUGCGAGAGUGAUCCUUCGAACCCUCU ----(((-(.....--(((-((..-.((((....(((.(((((....))))))))..(((.....((((((.....))))))...)))..)))).)))))((....))))))..... ( -37.80) >DroMoj_CAF1 3737 114 - 1 UAUAAUGGAGCUCAC-AAU-UUAU-UGCAGAUUCGGCUCGGGCAGCAGCCAAGCGUCACAGAACAGCGGCGAUAUAUGCCGCAUCUGCCACUGCGAGAGCGAUCCACAGAAUCCGCU .....((((((((..-...-...(-(((((.....(((..(((....))).)))....((((...((((((.....)))))).))))...))))))))))..))))........... ( -34.40) >DroAna_CAF1 2603 109 - 1 ----A---UUCUCUGAAAUAUCCU-UACAGGUUCGGUUCGGGCAGCAGCCAGGCCUCCCAGAACAGCGGCGACAUCUGCCGCAUCUGCCACUGCGAGAGCGAUCCCCAGAAUCCGCU ----.---...((((.........-..(((((...(((((((..((......))..))).)))).((((((.....)))))))))))....(((....))).....))))....... ( -32.30) >DroPer_CAF1 13509 108 - 1 ----AUU-CUCUGU--GCU-CUAU-UGCAGAUUCGGUUCUGGCAGCAGCCAGGCCUCGCAAAACAGCGGCGACAUCUGCCGCAUCUGCCACUGCGAGAGUGAUCCUUCGAACCCUCU ----(((-(((.((--(..-....-.((((((..(((.(((((....))))))))..........((((((.....)))))))))))))))...))))))................. ( -35.71) >consensus ____AUC_UGCUCU__AAU_CUAU_UGCAGAUUCGGUUCGGGCAGCAGCCAGGCCUCGCAGAACAGCGGCGACAUCUGCCGCAUCUGCCACUGCGAGAGCGAUCCACAGAAUCCGCU .............................(((((((.((.(((((..((...))...........((((((.....))))))..)))))...((....)))).))...))))).... (-25.80 = -25.80 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:08 2006