
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,357,302 – 9,357,437 |
| Length | 135 |
| Max. P | 0.590379 |

| Location | 9,357,302 – 9,357,414 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -24.59 |
| Energy contribution | -24.60 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9357302 112 - 23771897 UGCAAACAUUUUUGCAGUUGCUUUCGCCACUGCUUCGCGAAGUGGGA-AACGGGGCAAUUGAAAAUUUUGCAAAUCAUUGCCAGUCUGUUGCAAGUGUUUCGAUUAUUAUAUG ((((((....))))))(((.(((((.(((((.........)))))))-)).).)))((((((((..(((((((...((.....))...)))))))..))))))))........ ( -28.20) >DroSim_CAF1 7102 110 - 1 UGCAAACAUUUUUGCAGUUGCCUUCUCCACUGCUUCGCGAAGUGGGA-AACGGGGCAAUUGAAAAUUUUGCAAGUUAUUUGCUGUCUGUUGCAAG--UUUCGAUUAUUAUCUG ((((((...((((.((((((((((.((((((.........)))))).-...)))))))))))))).)))))).(..((((((........)))))--)..)(((....))).. ( -32.10) >DroEre_CAF1 6520 108 - 1 UGCAAACAUUUUUGCAGUUGCCUU-GCCACUGCUUCGCGAAGUGGGAAAACGGGGCAAUUGGAGAUUUUGCAAAUUAUUUGCAGUCUGUUGCAAG--AGUGGAUUAUUGUA-- (((((.((((((((((((((((((-(...(..(((....)))..).....))))))))))..((((..(((((.....)))))))))...)))))--)))).....)))))-- ( -34.80) >consensus UGCAAACAUUUUUGCAGUUGCCUUCGCCACUGCUUCGCGAAGUGGGA_AACGGGGCAAUUGAAAAUUUUGCAAAUUAUUUGCAGUCUGUUGCAAG__UUUCGAUUAUUAUAUG ((((((.(((((..((((((((((.....(..(((....)))..)......))))))))))))))))))))).....(((((........))))).................. (-24.59 = -24.60 + 0.01)

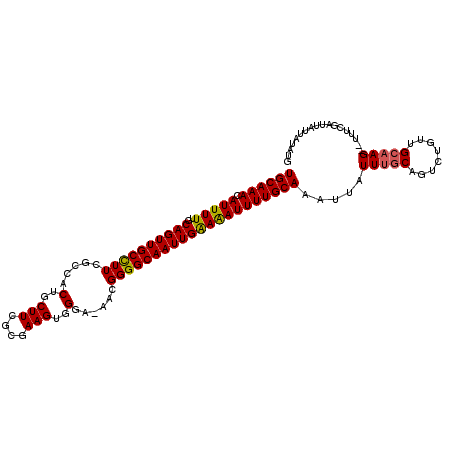
| Location | 9,357,342 – 9,357,437 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.52 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -16.86 |
| Energy contribution | -16.68 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9357342 95 - 23771897 AUGUUUGCCGUUCUUUUGUUUGUUGCAAACAUUUU------UGCAGUUGC-UUUCGCCACUGCUUCGCGAAGUGGGA-AACGGGGCAAUUGAAAAUUUUGCAA ((((((((((..(....)..))..))))))))(((------(.(((((((-((((.(((((.........)))))))-)....))))))))))))........ ( -24.40) >DroAna_CAF1 6583 103 - 1 GUGUUUGAGUUCUUUGUGUUUGCUGCAAACAUUUAGCCGCUUCUGGUGGCGCUCCUCUUUUACUUCCCGAAGUCAGGCACAAGGGCAAUUCAACGAUUUGCAA ((((((((.(((...(((((((...)))))))...(((((.....)))))..................))).))))))))....((((.((...)).)))).. ( -30.70) >DroSim_CAF1 7140 95 - 1 AUGUUUGCCGUUCUUUUGUUUGCUGCAAACAUUUU------UGCAGUUGC-CUUCUCCACUGCUUCGCGAAGUGGGA-AACGGGGCAAUUGAAAAUUUUGCAA ((((((((.((.(....)...)).))))))))(((------(.(((((((-(((.((((((.........)))))).-...))))))))))))))........ ( -28.30) >DroEre_CAF1 6556 95 - 1 AUGUUUGCCGUUCUUUUGUUUGCUGCAAACAUUUU------UGCAGUUGC-CUU-GCCACUGCUUCGCGAAGUGGGAAAACGGGGCAAUUGGAGAUUUUGCAA ((((((((.((.(....)...)).)))))))).((------(.(((((((-(((-(...(..(((....)))..).....))))))))))).)))........ ( -30.60) >consensus AUGUUUGCCGUUCUUUUGUUUGCUGCAAACAUUUU______UGCAGUUGC_CUUCGCCACUGCUUCGCGAAGUGGGA_AACGGGGCAAUUGAAAAUUUUGCAA ((((((((.((..........)).))))))))...........(((((((.((......((((((....))))))......)).)))))))............ (-16.86 = -16.68 + -0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:06 2006