
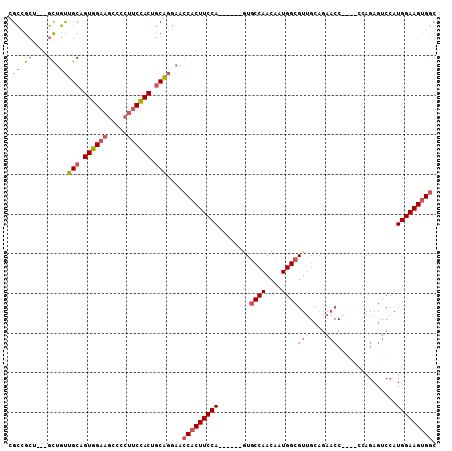
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,352,403 – 9,352,530 |
| Length | 127 |
| Max. P | 0.998176 |

| Location | 9,352,403 – 9,352,502 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.58 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -27.77 |
| Energy contribution | -30.28 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9352403 99 + 23771897 GCCACUUCCAUGGACUCUGAUUCUGGUUCUGCAACGCCAUUGUUGGCAC------UGGAAGUGGUUCCUGCGGUGGAAGGGGCUUCCACUGCAACAGC---AGCGGCA ((((((((((.(((((........)))))......((((....))))..------))))))))))...(((((((((((...)))))))))))...((---....)). ( -42.90) >DroAna_CAF1 1738 93 + 1 UCCUCUUCCAUGGGCUCUGG------------CACGCCAUUGUUGGAAGAAGAAGUGGAAGUGCUGGAAGUAGUAGUGGUGGCUUCCACGGCUGCAGC---CUCAACG ..........(((((((..(------------(((.(((((.((......)).)))))..))))..))....(((((.((((...)))).))))).))---).))... ( -33.90) >DroSim_CAF1 2353 99 + 1 GCCACUUCCAUGGACUCUGGUUCUGGUUCUGCAACGCCAUUGUUGGCAC------UGGAAGUGGUUCCUGCUGUGGAAGGGGCUUCCACUGCAACAGC---AGCAGCG ((((((((((.(((((........)))))......((((....))))..------))))))))))..((((((((((((...))))))).((....))---))))).. ( -40.90) >DroEre_CAF1 1858 96 + 1 GCCACUUCCAUUGACUCU------GGUUCUGCAACGCCAUUGUUGGCAC------UGGAAGUGGUUCCUGCAGUGGAAGGGGUUUCCACUGCAACAACAGCAGCGGCA (((((((((((..((..(------(((........))))..))..)...------))))))))))...(((((((((((...)))))))))))......((....)). ( -39.40) >consensus GCCACUUCCAUGGACUCUGG____GGUUCUGCAACGCCAUUGUUGGCAC______UGGAAGUGGUUCCUGCAGUGGAAGGGGCUUCCACUGCAACAGC___AGCAGCA ((((((((((.(((((........)))))......((((....))))........))))))))))...(((((((((((...)))))))))))............... (-27.77 = -30.28 + 2.50)



| Location | 9,352,403 – 9,352,502 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.58 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -21.42 |
| Energy contribution | -22.80 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9352403 99 - 23771897 UGCCGCU---GCUGUUGCAGUGGAAGCCCCUUCCACCGCAGGAACCACUUCCA------GUGCCAACAAUGGCGUUGCAGAACCAGAAUCAGAGUCCAUGGAAGUGGC ..(((((---((....)))))))..(((.((((((..(((((((....)))).------..((((....))))..))).((.......))........)))))).))) ( -35.80) >DroAna_CAF1 1738 93 - 1 CGUUGAG---GCUGCAGCCGUGGAAGCCACCACUACUACUUCCAGCACUUCCACUUCUUCUUCCAACAAUGGCGUG------------CCAGAGCCCAUGGAAGAGGA .((((.(---((....)))((((......)))).........)))).........((((((((((.....(((...------------.....)))..)))))))))) ( -30.40) >DroSim_CAF1 2353 99 - 1 CGCUGCU---GCUGUUGCAGUGGAAGCCCCUUCCACAGCAGGAACCACUUCCA------GUGCCAACAAUGGCGUUGCAGAACCAGAACCAGAGUCCAUGGAAGUGGC ..(((((---((....)).(((((((...))))))))))))...(((((((((------..((((....))))(((....)))...............))))))))). ( -37.60) >DroEre_CAF1 1858 96 - 1 UGCCGCUGCUGUUGUUGCAGUGGAAACCCCUUCCACUGCAGGAACCACUUCCA------GUGCCAACAAUGGCGUUGCAGAACC------AGAGUCAAUGGAAGUGGC .((((((((((((((((((((((((.....))))))))).((((....)))).------.....))))))))).........((------(.......))).)))))) ( -39.30) >consensus CGCCGCU___GCUGUUGCAGUGGAAGCCCCUUCCACUGCAGGAACCACUUCCA______GUGCCAACAAUGGCGUUGCAGAACC____CCAGAGUCCAUGGAAGUGGC .((.((....)).))(((.((((((.....)))))).)))....(((((((((........((((....))))(((....)))...............))))))))). (-21.42 = -22.80 + 1.38)


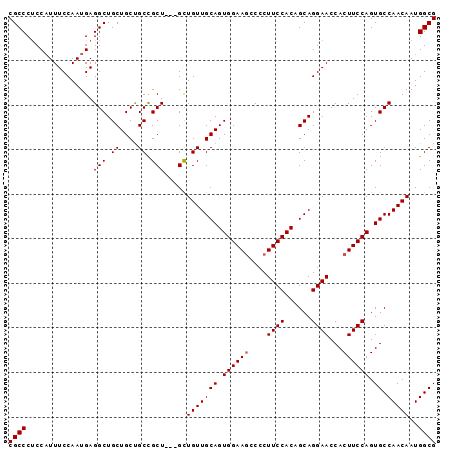
| Location | 9,352,437 – 9,352,530 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -28.63 |
| Energy contribution | -28.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9352437 93 - 23771897 CGCCCCCCAUUUCCAAUGAGGCUGCUGCUGCCGCU---GCUGUUGCAGUGGAAGCCCCUUCCACCGCAGGAACCACUUCCAGUGCCAACAAUGGCG .((..(((((.....))).))..))......((((---..(((((((.((((((....((((......))))...)))))).)).)))))..)))) ( -31.00) >DroSim_CAF1 2387 93 - 1 CGCCCUCCAUUUCCAAUGAGGCUGCUGCCGCUGCU---GCUGUUGCAGUGGAAGCCCCUUCCACAGCAGGAACCACUUCCAGUGCCAACAAUGGCG ((((......((((.....(((....)))((((((---((....))))((((((...)))))))))).)))).(((.....)))........)))) ( -31.50) >DroEre_CAF1 1886 96 - 1 CGCCCUCCCUUUCCAAUGAGGCUGCUGCUGCCGCUGCUGUUGUUGCAGUGGAAACCCCUUCCACUGCAGGAACCACUUCCAGUGCCAACAAUGGCG ((((...((((......))))....((.((.(((((..(((.(((((((((((.....))))))))))).)))......))))).)).))..)))) ( -34.60) >consensus CGCCCUCCAUUUCCAAUGAGGCUGCUGCUGCCGCU___GCUGUUGCAGUGGAAGCCCCUUCCACAGCAGGAACCACUUCCAGUGCCAACAAUGGCG ((((...............(((.((....)).))).....(((((((.((((((....((((......))))...)))))).)).)))))..)))) (-28.63 = -28.97 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:03 2006