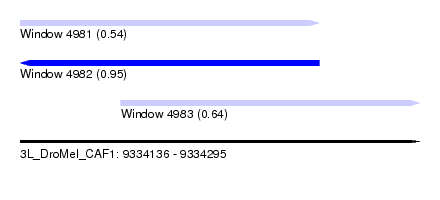
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,334,136 – 9,334,295 |
| Length | 159 |
| Max. P | 0.948860 |

| Location | 9,334,136 – 9,334,255 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9334136 119 + 23771897 UUUUGAUUUGAAAAGGAAUCGAUUUUUGGUGACGGCUUCAGAUCCA-UUUUCUCUAGCUACGUCGAUAGCAUGAACAGAUGAUUAUCCACAUCUGGUGGUUGCCUGUAUGGCCACCUGGU ....((((((((..(..(((((...)))))..)...))))))))..-.........((((......)))).....((((((........))))))..(((.(((.....))).))).... ( -29.20) >DroSec_CAF1 31608 115 + 1 U-UUGAUUUCAAAAGAAAUCGAUUCUUAGUGACGGAUCCAGAU-CA-UUUU--UUGGCUACGUCGAUAGCAUGAACAGAUGAAUAUCCACAUCUGGUGGUGGCCUGUAUGGCCACCUGGC .-((((((((....)))))))).......(((((...(((((.-..-...)--))))...)))))...((.....((((((........))))))..(((((((.....)))))))..)) ( -40.60) >DroSim_CAF1 31903 105 + 1 UUUUGAUUUCAAAAGAAAUCGAUUCUUAGUGACGGAUCCAGAUCCA-UUUU--UUGGCUACGUCGAUAGCAUGAACAGAUGAUUAUCCACAUCUG------------AUGGCCACCUGGU ..((((((((....))))))))..((...(((((...(((((....-...)--))))...)))))..))......((((((........))))))------------...(((....))) ( -26.10) >DroEre_CAF1 26232 105 + 1 UUUUAAUUUGAAAAGAAAUCGAUUUUAAGUGACGGAUCCAGAUCCA-UUUU--CUGGCUAUGUCGAUAGCUCGAACAGAUGAUUAUCCACAUCUGAUG------------GCCACCUGGU .....((((((((.(....)..))))))))...((((....)))).-....--.(((((((.((((....)))).((((((........)))))))))------------))))...... ( -25.00) >DroYak_CAF1 37835 118 + 1 CUUUGAUUUCAAAAGAAAUCGAUUUCCAGUGACGGAUCCAGAUCCGCUUUU--GUGGCUACGUCGAUAGCUCGAACAGAUGAUUAUCCACAUCUGGUGGCUGCCUGUUUGGCCACCUGGU ..((((((((....))))))))...((((...(((((....))))).....--(((((((..(.(.(((((....((((((........))))))..))))).).)..))))))))))). ( -39.80) >consensus UUUUGAUUUCAAAAGAAAUCGAUUUUUAGUGACGGAUCCAGAUCCA_UUUU__CUGGCUACGUCGAUAGCAUGAACAGAUGAUUAUCCACAUCUGGUGG__GCCUGUAUGGCCACCUGGU ..((((((((....)))))))).......(((((...((((............))))...)))))..........((((((........))))))...............(((....))) (-19.72 = -19.92 + 0.20)

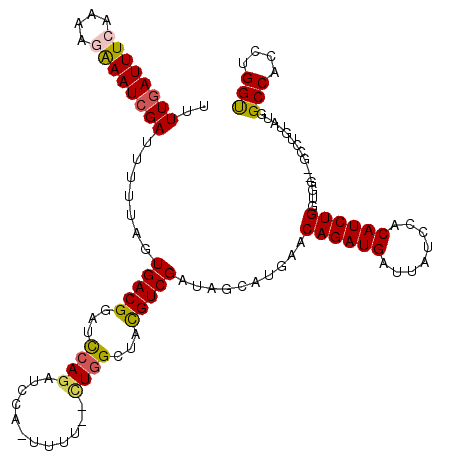
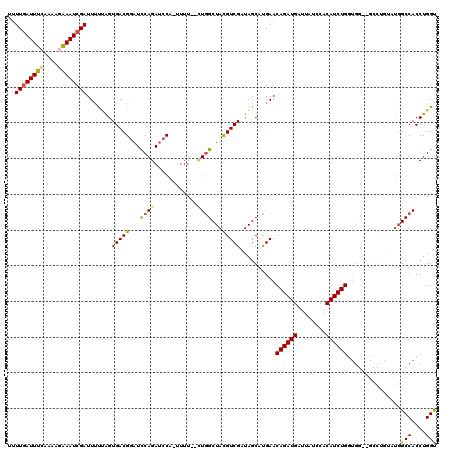
| Location | 9,334,136 – 9,334,255 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9334136 119 - 23771897 ACCAGGUGGCCAUACAGGCAACCACCAGAUGUGGAUAAUCAUCUGUUCAUGCUAUCGACGUAGCUAGAGAAAA-UGGAUCUGAAGCCGUCACCAAAAAUCGAUUCCUUUUCAAAUCAAAA ..(((((((((.....)))..((((.....)))).....)))))).((..(((((....)))))..))(((((-.(((..(((...............)))..))))))))......... ( -26.46) >DroSec_CAF1 31608 115 - 1 GCCAGGUGGCCAUACAGGCCACCACCAGAUGUGGAUAUUCAUCUGUUCAUGCUAUCGACGUAGCCAA--AAAA-UG-AUCUGGAUCCGUCACUAAGAAUCGAUUUCUUUUGAAAUCAA-A ....(((((((.....))))))).(((((((((((((......)))))))(((((....)))))...--....-..-)))))).................((((((....))))))..-. ( -37.60) >DroSim_CAF1 31903 105 - 1 ACCAGGUGGCCAU------------CAGAUGUGGAUAAUCAUCUGUUCAUGCUAUCGACGUAGCCAA--AAAA-UGGAUCUGGAUCCGUCACUAAGAAUCGAUUUCUUUUGAAAUCAAAA ....((((((...------------((((((........)))))).....))))))((((...(((.--....-......)))...))))..........((((((....)))))).... ( -27.90) >DroEre_CAF1 26232 105 - 1 ACCAGGUGGC------------CAUCAGAUGUGGAUAAUCAUCUGUUCGAGCUAUCGACAUAGCCAG--AAAA-UGGAUCUGGAUCCGUCACUUAAAAUCGAUUUCUUUUCAAAUUAAAA ....((((((------------(..((((((........))))))...).))))))(((....((((--(...-....)))))....))).............................. ( -24.60) >DroYak_CAF1 37835 118 - 1 ACCAGGUGGCCAAACAGGCAGCCACCAGAUGUGGAUAAUCAUCUGUUCGAGCUAUCGACGUAGCCAC--AAAAGCGGAUCUGGAUCCGUCACUGGAAAUCGAUUUCUUUUGAAAUCAAAG .(((((((((......(..((((..((((((........))))))...).)))..)......)))))--....((((((....))))))..)))).....((((((....)))))).... ( -39.70) >consensus ACCAGGUGGCCAUACAGGC__CCACCAGAUGUGGAUAAUCAUCUGUUCAUGCUAUCGACGUAGCCAA__AAAA_UGGAUCUGGAUCCGUCACUAAAAAUCGAUUUCUUUUGAAAUCAAAA ....((((((...............((((((........)))))).....(((((....)))))...........((((....)))))))))).......((((((....)))))).... (-22.58 = -23.10 + 0.52)



| Location | 9,334,176 – 9,334,295 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.53 |
| Mean single sequence MFE | -39.84 |
| Consensus MFE | -26.80 |
| Energy contribution | -27.76 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9334176 119 + 23771897 GAUCCA-UUUUCUCUAGCUACGUCGAUAGCAUGAACAGAUGAUUAUCCACAUCUGGUGGUUGCCUGUAUGGCCACCUGGUAACACUGCACCAUGGGGGUGGUGAUAAAAGGGGGUGUUGU .((((.-.......(((((((((.....)).....((((((........)))))))))))))(((.(((.((((((((((........)))....))))))).)))..)))))))..... ( -34.80) >DroSec_CAF1 31647 116 + 1 GAU-CA-UUUU--UUGGCUACGUCGAUAGCAUGAACAGAUGAAUAUCCACAUCUGGUGGUGGCCUGUAUGGCCACCUGGCAACACUGCACCAUGGUGGUGGUGAUAAAAGGGGGUGUUGU ..(-((-(((.--(((((...))))).)).)))).((((((........))))))..(((((((.....)))))))..((((((((.((((((....)))))).........)))))))) ( -40.80) >DroSim_CAF1 31943 105 + 1 GAUCCA-UUUU--UUGGCUACGUCGAUAGCAUGAACAGAUGAUUAUCCACAUCUG------------AUGGCCACCUGGUAACACUGCACCAUGGGGGUGGUGAUAAAAGGGGGUGUUGU .((((.-....--.((((((((((....).)))..((((((........))))))------------.))))))(((..((.(((..(.((....)))..))).))..)))))))..... ( -32.40) >DroEre_CAF1 26272 105 + 1 GAUCCA-UUUU--CUGGCUAUGUCGAUAGCUCGAACAGAUGAUUAUCCACAUCUGAUG------------GCCACCUGGUAACACUGCCCCAUGGGGGUGGUGAUACAAGGGGGUGUUGU .((((.-....--.(((((((.((((....)))).((((((........)))))))))------------))))(((.(((.(((..((((...))))..))).))).)))))))..... ( -43.70) >DroYak_CAF1 37875 118 + 1 GAUCCGCUUUU--GUGGCUACGUCGAUAGCUCGAACAGAUGAUUAUCCACAUCUGGUGGCUGCCUGUUUGGCCACCUGGUAACACUGCCCCAUGGGGGUGGUGAUAAAAGGGGGUGUUGU .((((.(((((--((((((((.((((....)))).((((((........))))))))))))(((.....)))..........(((..((((...))))..)))))))))).))))..... ( -47.50) >consensus GAUCCA_UUUU__CUGGCUACGUCGAUAGCAUGAACAGAUGAUUAUCCACAUCUGGUGG__GCCUGUAUGGCCACCUGGUAACACUGCACCAUGGGGGUGGUGAUAAAAGGGGGUGUUGU .((((.........((((((..(((......))).((((((........)))))).............))))))(((..((.(((..(.((....)))..))).))..)))))))..... (-26.80 = -27.76 + 0.96)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:53 2006