
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,311,465 – 9,311,583 |
| Length | 118 |
| Max. P | 0.769139 |

| Location | 9,311,465 – 9,311,583 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.28 |
| Mean single sequence MFE | -45.33 |
| Consensus MFE | -28.00 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9311465 118 + 23771897 GACAUUGGCCAUUACCUUUAUGGGCGACUAUGGCCGGUUUAAGCCCGGUCCCAAACAGCUGGAAGGUGUCCAAUUUCUGCUGGCCCAUGCAGUGGCCAAUCGAAAUAUUGAUGUGGAC ..(((((((((((.....(((((((......((((((.......)))))).....((((.(((((........)))))))))))))))).))))))))(((((....))))))))... ( -41.70) >DroVir_CAF1 24488 118 + 1 CACACUGGCCAUCACCUUUAUGGGCGACUAUGGCCGCUAUCAGCCCACCGCCAAGCAGCUGGAGGGCGUGCAAUUCCUUUUGGCCCAUGCCGUGGCCAAUCACAAAUUGGAUUUGGAC ..(((.(((((((.(((....))).))...))))).......((((.((((......)).)).)))))))(((.(((..(((((((.....).)))))).........))).)))... ( -40.00) >DroGri_CAF1 25625 118 + 1 CACACUGGCUAUUACCUUUAUGGGUGACUAUGCCCGCUACACGCCCACCCCCAAGCAGCUGGAGGGAGUUCAGUUCCUCCUGGCCCAUGGGGUGGCCAAUCGCAAGCUGCAUGUGGAC ......(((..((((((....))))))....)))..(((((.(((((((((......((((((((((......))))))).)))....))))))).....(....)..)).))))).. ( -43.40) >DroYak_CAF1 10735 118 + 1 GACAUUGGCCAUUACCUUCAUGGGCGACUACGGUCGCUACAAGCCCAGUGCCAAGCAGCUGGAAGGUGUCCAAUUCCUGUUGGCCCAUGCCGUGGCCAACAGGAAUCUUGAGGUGGAC ....((.(((..(((((((...((((((....)))))).......((((((...))).))))))))))....((((((((((((((.....).))))))))))))).....))).)). ( -52.60) >DroMoj_CAF1 19464 118 + 1 CACACUCGCCAUCACAUUUAUGGGCGACUAUGGCCGCUAUGUGCCCACACCCAAGCAGCUGGAGGGCGUGCAGUUCCUGCUGGCCCAUGCCGUGGCCAAUCGCAAAGUGGAUUUAGAC ..((((..((((.......))))((((...(((((((.....((((....(((......))).))))..((((...)))).(((....)))))))))).))))..))))......... ( -39.50) >DroAna_CAF1 2574 118 + 1 GACACUGGCCGUUACAUUUAUGGGCGACUACGGUCGCUUCCAGCCCUCCGCCAAACAGCUGGAGGGCGUCCAAUUUCUGUUGGCCCAUGCCGUGGCCAAUGGAGCCUUGGAGGUGGAU ..((((((((((.......))))(((((....)))))..)).(((((((((......)).))))))).(((((.((((((((((((.....).)))))))))))..)))))))))... ( -54.80) >consensus CACACUGGCCAUUACCUUUAUGGGCGACUAUGGCCGCUACAAGCCCACCCCCAAGCAGCUGGAGGGCGUCCAAUUCCUGCUGGCCCAUGCCGUGGCCAAUCGCAAACUGGAUGUGGAC .....(((((((......(((((((......(((........)))..........((((.((((.........)))).)))))))))))..))))))).................... (-28.00 = -27.20 + -0.80)
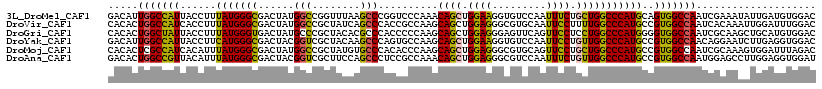

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:50 2006