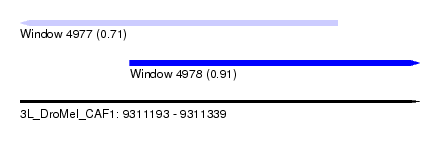
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,311,193 – 9,311,339 |
| Length | 146 |
| Max. P | 0.908520 |

| Location | 9,311,193 – 9,311,309 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -21.58 |
| Energy contribution | -23.26 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9311193 116 - 23771897 CACGCGCCGGGAUUUCUUUUCCCUACUACACUCGAAGAAAUUUUGAUUUUUAAGGCUUAAAAAGUAUUAUACAAGGCUUAUUUUAGGAACUAUGUUUCCCAUUGAGGCGCGUAAAA .(((((((((((.......))))........((((.(((((...(.((((((((((((...............)))))....))))))))...)))))...))))))))))).... ( -28.06) >DroSec_CAF1 9245 115 - 1 CAAGCGCUGGGACUUCUUUACCCUACUACACUCGAAGAAAUUUGUAU-GUUACAGCUUUAAAAAAGUUAUAUAAGGCUUACUUUAGGGACUAUGUUUCCCAUUAAGGCGUGUAAAA ...(((((((((........(((((..(((..((((....))))..)-))...(((((((...........))))))).....)))))........)))).....)))))...... ( -25.59) >DroSim_CAF1 9347 115 - 1 CACGCUCUGGGAUUUCUUUUCCCUACUACACUCGAAGAAAUUUGUAU-GUUAAAGCUUUAAAAUAGUUAUAUAAGGCUUAUUUUAGGGACUAUGUUUCCCAUUAAGGCGUGUAAAA ((((((.(((((.......((((((..(((..((((....))))..)-))..((((((((...........))))))))....)))))).......)))))....))))))..... ( -33.44) >consensus CACGCGCUGGGAUUUCUUUUCCCUACUACACUCGAAGAAAUUUGUAU_GUUAAAGCUUUAAAAAAGUUAUAUAAGGCUUAUUUUAGGGACUAUGUUUCCCAUUAAGGCGUGUAAAA .(((((((((((........(((((.......((((....))))........((((((((...........))))))))....)))))........)))).....))))))).... (-21.58 = -23.26 + 1.67)

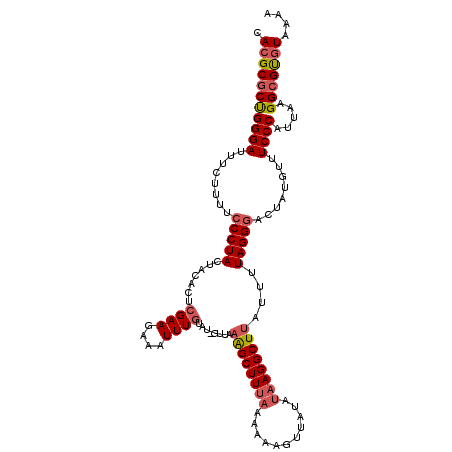
| Location | 9,311,233 – 9,311,339 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.36 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -19.22 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9311233 106 + 23771897 CCUUGUAUAAUACUUUUUAAGCCUUAAAAAUCAAAAUUUCUUCGAGUGUAGUAGGGAAAAGAAAUCCCGGCGCGUGCAUUUGUGUUCCCAUCACGUUCACGUGCCA .....(((.((((((............................)))))).)))((((.......))))((((((((....((((.......))))..)))))))). ( -24.89) >DroSec_CAF1 9285 105 + 1 CCUUAUAUAACUUUUUUAAAGCUGUAAC-AUACAAAUUUCUUCGAGUGUAGUAGGGUAAAGAAGUCCCAGCGCUUGCAUUUGUGUUCCCGGCACGUUCACGUGCCA .....((((.(((.....))).))))..-((((((((.....(((((((....(((.........))).))))))).))))))))....((((((....)))))). ( -26.30) >DroSim_CAF1 9387 105 + 1 CCUUAUAUAACUAUUUUAAAGCUUUAAC-AUACAAAUUUCUUCGAGUGUAGUAGGGAAAAGAAAUCCCAGAGCGUGCAUUUGUGUUCCCGGCACGUUCACGUGCCA ............................-.............((((((((((.((((.......))))...)).)))))))).......((((((....)))))). ( -23.50) >DroEre_CAF1 2653 94 + 1 ---------AUAUUUUUUAAGCUCCAAC-AAAGAAAUUUGUUCGAGUGUAGUAGGAAAAGAAA--UCCGGCGCGUGCAUUUGUGUUCCCGGCACGUUCACGUGCCA ---------...(((((((.((((.(((-(((....)))))).))))....))))))).....--...((((((((....(((((.....)))))..)))))))). ( -33.50) >DroYak_CAF1 10519 90 + 1 --------------UUAGAAGCCCC-UU-AAGGCAUUUUUUUCGAGUGUAGUAGGAAGUGAAAAUCCCGGCGCGUGCAUUUGUGUUCCCGGCACGUUCACGUGCCU --------------......((..(-((-(..((((((.....))))))..))))..)).........((((((((....(((((.....)))))..)))))))). ( -26.90) >consensus CCUU_UAUAACAUUUUUAAAGCUCUAAC_AUACAAAUUUCUUCGAGUGUAGUAGGGAAAAGAAAUCCCGGCGCGUGCAUUUGUGUUCCCGGCACGUUCACGUGCCA ..........................................((((((((((.((((.......))))...)).)))))))).......((((((....)))))). (-19.22 = -19.98 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:48 2006