| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
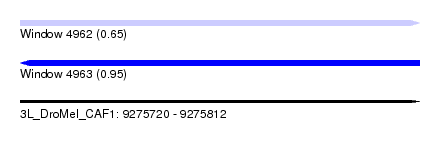
| Location | 9,275,720 – 9,275,812 |
| Length | 92 |
| Max. P | 0.950524 |

| Location | 9,275,720 – 9,275,812 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -11.61 |
| Energy contribution | -11.92 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
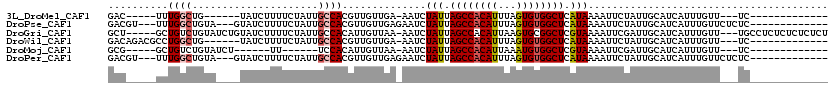
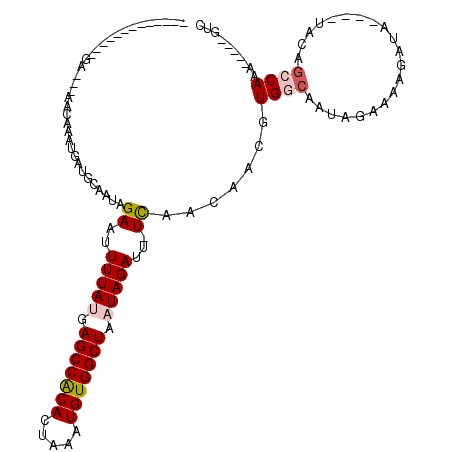
>3L_DroMel_CAF1 9275720 92 + 23771897 GAC-----UUUGGCUG------UAUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUU---UC------------- ...-----..((((.(------((.......))).)))).((.((.((-(.(.(((.((((((((...)))))))).))).).))).)).))..........---..------------- ( -19.10) >DroPse_CAF1 134803 101 + 1 GACGU---UUUGGCUGUA---GUAUCUUUUCUAUUGCCACGUUGUUGAGAAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUCUCUC------------- (((((---...(((.(((---(........)))).))))))))((..(((((.(((.((((((((...)))))))).)))..)))))...))...............------------- ( -27.00) >DroGri_CAF1 124218 111 + 1 GCU-----GCUGUCUGUAUCUGUAUCUUUUCUAUUGCCACAUUGUUAA-AAUCUAUUAGCCACAUUAAGUGCGGCUCGUAAAAUUCGAUUGCAUCAUUUGUU---UGCCUCUCUCUCUCU (.(-----((.(((.......(((..........)))...........-....(((.((((.(((...))).)))).)))......))).))).).......---............... ( -12.50) >DroWil_CAF1 213739 97 + 1 GACAGACGCCUGGCUG------UAUCUUUUCUAUUGCCACGUUGUUGA-AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUU---UC------------- ((((((.((.((((.(------((.......))).)))).))(((.((-(.(.(((.((((((((...)))))))).))).).)))....)))...))))))---..------------- ( -22.20) >DroMoj_CAF1 143823 86 + 1 GCG-----GCUGUCUGUAUCU------UU------UCCACAUUGUUAA-AAUCUAUUAGCCACAUUAAAUGUGGCUCGUAAAAUUCGAUUGCAUCAUUUGUU---UC------------- ...-----((.((((((....------..------...))).......-....(((.((((((((...)))))))).)))......))).))..........---..------------- ( -14.60) >DroPer_CAF1 135765 101 + 1 GACGU---UUUGGCUGUA---GUAUCUUUUCUAUUGCCACGUUGUUGAGAAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUUCUCUC------------- (((((---...(((.(((---(........)))).))))))))((..(((((.(((.((((((((...)))))))).)))..)))))...))...............------------- ( -27.00) >consensus GAC_____UCUGGCUGUA____UAUCUUUUCUAUUGCCACGUUGUUGA_AAUCUAUUAGCCACAUUUAGUGUGGCUCAUAAAAUUCUAUUGCAUCAUUUGUU___UC_____________ ..........((((.....................))))..............(((.((((((((...)))))))).)))........................................ (-11.61 = -11.92 + 0.31)
| Location | 9,275,720 – 9,275,812 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.85 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
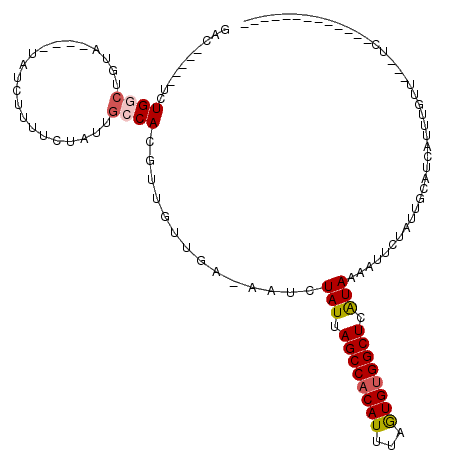
>3L_DroMel_CAF1 9275720 92 - 23771897 -------------GA---AACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUA------CAGCCAAA-----GUC -------------..---................(((.(((((.(((((((.....))))))).))))).)-)).......((((..((.......))------..))))..-----... ( -20.30) >DroPse_CAF1 134803 101 - 1 -------------GAGAGAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUUCUCAACAACGUGGCAAUAGAAAAGAUAC---UACAGCCAAA---ACGUC -------------............((((....(((((((....(((((((.....)))))))...)))))))........((((..(((........)---))..))))..---.)))) ( -23.50) >DroGri_CAF1 124218 111 - 1 AGAGAGAGAGAGGCA---AACAAAUGAUGCAAUCGAAUUUUACGAGCCGCACUUAAUGUGGCUAAUAGAUU-UUAACAAUGUGGCAAUAGAAAAGAUACAGAUACAGACAGC-----AGC ...............---.........(((..(((((.((((..(((((((.....)))))))..)))).)-)).....((((.............))))......))..))-----).. ( -16.52) >DroWil_CAF1 213739 97 - 1 -------------GA---AACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU-UCAACAACGUGGCAAUAGAAAAGAUA------CAGCCAGGCGUCUGUC -------------..---.......(((((....(((.(((((.(((((((.....))))))).))))).)-)).......((((..((.......))------..)))).))))).... ( -27.20) >DroMoj_CAF1 143823 86 - 1 -------------GA---AACAAAUGAUGCAAUCGAAUUUUACGAGCCACAUUUAAUGUGGCUAAUAGAUU-UUAACAAUGUGGA------AA------AGAUACAGACAGC-----CGC -------------..---..........((..(((((.((((..((((((((...))))))))..)))).)-)).....((((..------..------...))))))..))-----... ( -16.30) >DroPer_CAF1 135765 101 - 1 -------------GAGAGAACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUUCUCAACAACGUGGCAAUAGAAAAGAUAC---UACAGCCAAA---ACGUC -------------............((((....(((((((....(((((((.....)))))))...)))))))........((((..(((........)---))..))))..---.)))) ( -23.50) >consensus _____________GA___AACAAAUGAUGCAAUAGAAUUUUAUGAGCCACACUAAAUGUGGCUAAUAGAUU_UCAACAACGUGGCAAUAGAAAAGAUA____UACAGCCAAA_____GUC ..................................((..(((((.(((((((.....))))))).)))))...)).......((((.....................)))).......... (-13.38 = -13.85 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:34 2006