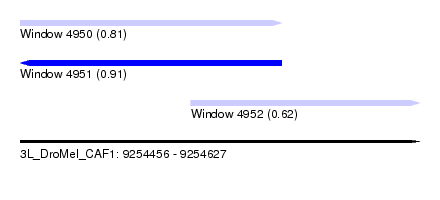
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,254,456 – 9,254,627 |
| Length | 171 |
| Max. P | 0.907843 |

| Location | 9,254,456 – 9,254,568 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -30.64 |
| Consensus MFE | -16.67 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
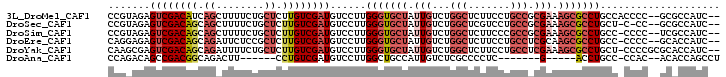
>3L_DroMel_CAF1 9254456 112 + 23771897 CUACGAGUAUUA-----U-UAUGCAACCAA-UUUUUUUUUCUCCGUCCUGUUGCUGCAACACCAACGGUGGCAAAUGUUGCCGUAGAGUCGACAUCAGCUUUUCUGCUCUUGUCGAUGU (((((.((((..-----.-.))))...(((-(...(((.((.((((..(((((...)))))...)))).)).))).)))).))))).(((((((..(((......)))..))))))).. ( -31.50) >DroSec_CAF1 101271 111 + 1 CUACGAGUACUA-----U-UUUGCAACC-A-UUUUUUUUUCUCCGUCCUGUUGCUGCAACACCAACGGUGGCAAAUGUUGCCGUAGAGUCGACAGCAGCUUUUCUGCUCUUGUCGAUGU (((((.(((.((-----(-((.((....-.-...........((((..(((((...)))))...))))..))))))).)))))))).((((((((.(((......))).)))))))).. ( -31.87) >DroSim_CAF1 101646 113 + 1 CUACGAGUACUA-----U-UUUGCAACCCAUUUUUUUUUUCUCCGUUCUGUUGCUGCAACACCAACGGUGGCAAAUGUUGCCGUAGAGUCGACAGCAGCUUUUCUGCUCUUGUCGAUGU (((((.(((.((-----(-((.((..................(((((.(((((...)))))..)))))..))))))).)))))))).((((((((.(((......))).)))))))).. ( -32.35) >DroEre_CAF1 124047 106 + 1 CCACGAGUAAUC-----U-GCUACAACCUA-----UUUUUCUCCGUGC--UUGCUGCAACACCAACGGUAGCAAAUGUUGCAGGAGAGUCGACAGCAGAUUCUCCGCUCUUGUCGAUGU .((.((((((((-----(-(((........-----....(((((.(((--(((((((..........))))))).....))))))))(....)))))))))....)))).))....... ( -29.40) >DroYak_CAF1 118927 108 + 1 CUAUGAGUAAUC-----U-UUUGCCACCAA-----UUUUUCUCCGUCCAGUUGCUGCAACAACAGCGGUGGCAAAUGUUGCAAGCGAGUCGACAGCAGAUUUUCUGCUCUUGUCGAUGU ......(((((.-----.-(((((((((..-----........(.....)..((((......))))))))))))).)))))......(((((((((((.....))))...))))))).. ( -36.80) >DroAna_CAF1 101990 108 + 1 UUGGGAAUAAACAGCAAUUUUUAC-AUAUA---UUUUUUUGCCUGUUC-CUUGAAGCAAUAAAAACAGUGGCAACUGUUGCCAGACAGCCGACGGCAGACUU------CCUGUCGAUGU ..(((((((....((((.......-.....---.....)))).)))))-))....(((..........(((((.....))))).........((((((....------.)))))).))) ( -21.93) >consensus CUACGAGUAAUA_____U_UUUGCAACCAA___UUUUUUUCUCCGUCCUGUUGCUGCAACACCAACGGUGGCAAAUGUUGCCGGAGAGUCGACAGCAGAUUUUCUGCUCUUGUCGAUGU ..........................................((((...((((...))))....))))(((((.....)))))....((((((((.((........)).)))))))).. (-16.67 = -17.12 + 0.45)


| Location | 9,254,456 – 9,254,568 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -31.81 |
| Consensus MFE | -17.80 |
| Energy contribution | -19.25 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9254456 112 - 23771897 ACAUCGACAAGAGCAGAAAAGCUGAUGUCGACUCUACGGCAACAUUUGCCACCGUUGGUGUUGCAGCAACAGGACGGAGAAAAAAAAA-UUGGUUGCAUA-A-----UAAUACUCGUAG ...((((((..(((......)))..))))))..((((((((((((..((....))..))))))).(((((..................-...)))))...-.-----.......))))) ( -31.50) >DroSec_CAF1 101271 111 - 1 ACAUCGACAAGAGCAGAAAAGCUGCUGUCGACUCUACGGCAACAUUUGCCACCGUUGGUGUUGCAGCAACAGGACGGAGAAAAAAAAA-U-GGUUGCAAA-A-----UAGUACUCGUAG ...((((((..(((......)))..))))))..((((((((((((..((....))..))))))).(((((..................-.-.)))))...-.-----.......))))) ( -31.65) >DroSim_CAF1 101646 113 - 1 ACAUCGACAAGAGCAGAAAAGCUGCUGUCGACUCUACGGCAACAUUUGCCACCGUUGGUGUUGCAGCAACAGAACGGAGAAAAAAAAAAUGGGUUGCAAA-A-----UAGUACUCGUAG ...((((((..(((......)))..))))))..((((((((((((..((....))..))))))).(((((....((.............)).)))))...-.-----.......))))) ( -31.92) >DroEre_CAF1 124047 106 - 1 ACAUCGACAAGAGCGGAGAAUCUGCUGUCGACUCUCCUGCAACAUUUGCUACCGUUGGUGUUGCAGCAA--GCACGGAGAAAAA-----UAGGUUGUAGC-A-----GAUUACUCGUGG ............((((..(((((((((.(((((((((((((((((..((....))..))))))))(...--.)..)))))....-----...))))))))-)-----))))..)))).. ( -36.71) >DroYak_CAF1 118927 108 - 1 ACAUCGACAAGAGCAGAAAAUCUGCUGUCGACUCGCUUGCAACAUUUGCCACCGCUGUUGUUGCAGCAACUGGACGGAGAAAAA-----UUGGUGGCAAA-A-----GAUUACUCAUAG ...((((((...((((.....)))))))))).............((((((((((((((....)))))..((....)).......-----..)))))))))-.-----............ ( -34.70) >DroAna_CAF1 101990 108 - 1 ACAUCGACAGG------AAGUCUGCCGUCGGCUGUCUGGCAACAGUUGCCACUGUUUUUAUUGCUUCAAG-GAACAGGCAAAAAAA---UAUAU-GUAAAAAUUGCUGUUUAUUCCCAA .....(((((.------((...(((....(((...(((....)))..)))..(((((((.((((((....-....)))))))))))---))...-)))....)).)))))......... ( -24.40) >consensus ACAUCGACAAGAGCAGAAAAGCUGCUGUCGACUCUACGGCAACAUUUGCCACCGUUGGUGUUGCAGCAACAGGACGGAGAAAAAAA___UAGGUUGCAAA_A_____GAUUACUCGUAG ...((((((...((((.....)))))))))).......(((((((..((....))..))))))).(((((......................)))))...................... (-17.80 = -19.25 + 1.45)

| Location | 9,254,529 – 9,254,627 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -19.77 |
| Energy contribution | -21.72 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9254529 98 + 23771897 CCGUAGAGUCGACAUCAGCUUUUCUGCUCUUGUCGAUGUCCUUGGGUGCUAUUGUCUGGCUCUUCCUGCCGCGAAAGCGCCUGCCACCCC--GCGCCAUC-- .((..(.(((((((..(((......)))..)))))))......(((((((.((((..(((.......))))))).)))))))......).--.)).....-- ( -33.80) >DroSec_CAF1 101343 96 + 1 CCGUAGAGUCGACAGCAGCUUUUCUGCUCUUGUCGAUGUCCUUGGGUGCUAUUGUCUGGCUCGUCCUGCCGCGAAAGCGCCUGCU-C-CC--GCGCCAUC-- .(((.((((((((((.(((......))).))))))........(((((((.((((..(((.......))))))).))))))))))-)-..--))).....-- ( -34.80) >DroSim_CAF1 101720 97 + 1 CCGUAGAGUCGACAGCAGCUUUUCUGCUCUUGUCGAUGUCCUUGGGUGCUAUUGUCUGGCUCUUCCCGCCGCGAAAGCGCCUGCC-CCCC--UCGCCAUC-- .....((((((((((.(((......))).))))))).......(((((((.((((..(((.......))))))).)))))))...-...)--))......-- ( -33.10) >DroEre_CAF1 124114 97 + 1 CAGGAGAGUCGACAGCAGAUUCUCCGCUCUUGUCGAUGUCCUUGGGUGCUAUUGUCUGGCUCUUCCUGCCUCGCAAGCGCCUGCC-CCCC--GCACCAUC-- .((((..((((((((.((........)).)))))))).)))).(((((((..(((..(((.......)))..))))))))))((.-....--))......-- ( -33.00) >DroYak_CAF1 118996 99 + 1 CAAGCGAGUCGACAGCAGAUUUUCUGCUCUUGUCGAUGUCCUUGGGUGCUAUUGUCUGGCUCUUCCUGCCUCGAAAGCGCCUGCU-CCCCGCGCACCAUC-- ...(((.(((((((((((.....))))...)))))))......(((((((.(((...(((.......))).))).)))))))...-...)))........-- ( -36.00) >DroAna_CAF1 102065 81 + 1 CCAGACAGCCGACGGCAGACUU------CCUGUCGAUGUCCUUGGCUGCCAUUGUCUCGCCCCUC-------G-----ACCUGCC-CCAC--ACACCAGCCU ...((((..((((((.......------.)))))).))))...(((((....(((...((.....-------.-----....)).-..))--)...))))). ( -19.90) >consensus CCGGAGAGUCGACAGCAGAUUUUCUGCUCUUGUCGAUGUCCUUGGGUGCUAUUGUCUGGCUCUUCCUGCCGCGAAAGCGCCUGCC_CCCC__GCACCAUC__ .......((((((((.((........)).))))))))......(((((((.(((...(((.......))).))).))))))).................... (-19.77 = -21.72 + 1.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:23 2006