
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
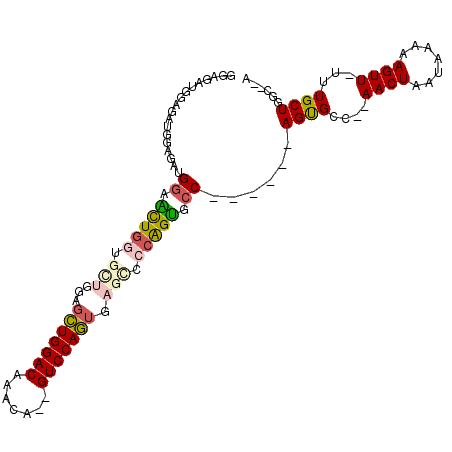
| Location | 9,205,683 – 9,205,780 |
| Length | 97 |
| Max. P | 0.977282 |

| Location | 9,205,683 – 9,205,780 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9205683 97 + 23771897 GGAGAUGGUGAUGGAGAUGGAACUGGUGCUCGAGCUGGACAAACA--GUCCAGUGAGCCCCAGUGCC------AGUGCC--AACUAAUAAAAAGUUUUUUGCUGGC--A .....((((..(((...(((.(((((.((((..(((((((.....--))))))))))).))))).))------)...))--)))))......(((.....)))...--. ( -35.90) >DroSec_CAF1 59473 97 + 1 GGUGAUGGUGAUGGAGAUGGAACUGCUGCUGGAGCUGGACAAACA--GUCCAGUGAGCCCCAGUGCC------AGUGCC--AACUAAUAAAAAGUUUUUUGCUGGC--A .((..((((..(((...(((.((((..(((...(((((((.....--))))))).)))..)))).))------)...))--)))))......(((.....))).))--. ( -30.00) >DroSim_CAF1 60030 97 + 1 GGAGAUGGUGAUGGAGAUGGAACUGGUGCUGGAGCUGGACAAACA--GUCCAGUGAGCCCCAGUGCC------AGUGCC--AACUAAUAAAAAGUUUUUUGCUGGC--A ((((((..(.....((.(((.(((((..((((.(((((((.....--))))))).....))))..))------))).))--).)).....)..)))))).......--. ( -36.80) >DroEre_CAF1 77174 96 + 1 GGAGAUGGAGAUGGAGAUGGAGCUGGU------GCUGGACAAACA--GUCCAGUGAGCCCCAGUGCCAGUGCCAGUGCC--AACUAAUAAAAAGUU-UUUGCUGGC--A ...........(((...(((.(((..(------(((((((.....--))))))))))).)))...))).((((((((..--((((.......))))-..)))))))--) ( -34.70) >DroYak_CAF1 76339 94 + 1 --GGAUGUAAAUGGAGAUGGAGUUGGU------GCUGGACAAAUA--GUCCAGUGAGCCCCAGUGCCAGUGCCAGUGCC--AACUAAUAAAAAGUU-UUUGCUGGC--A --.........(((...(((.((...(------(((((((.....--)))))))).)).)))...))).((((((((..--((((.......))))-..)))))))--) ( -32.50) >DroPer_CAF1 69735 99 + 1 GAAGCUAGAGUUUGAGAUGGAGUUGAAGUUGCAGAUGGACAAACAGAGUCCAU--AGUCCGGGCAGC------AGCGUCGAAACUAAUAAAAAGUU--UUGCUGGCACA ...(((((...................(((((..((((((.......))))))--.((....)).))------)))..(((((((.......))))--))))))))... ( -26.70) >consensus GGAGAUGGAGAUGGAGAUGGAACUGGUGCUGGAGCUGGACAAACA__GUCCAGUGAGCCCCAGUGCC______AGUGCC__AACUAAUAAAAAGUU_UUUGCUGGC__A ..................((.(((((.(((...(((((((.......))))))).))).))))).))......((((....((((.......))))...))))...... (-17.96 = -18.47 + 0.50)


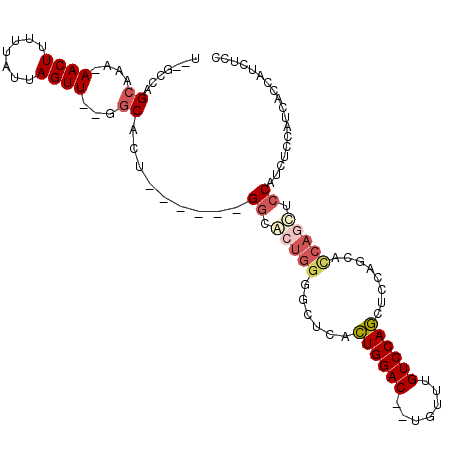
| Location | 9,205,683 – 9,205,780 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -14.91 |
| Energy contribution | -15.86 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9205683 97 - 23771897 U--GCCAGCAAAAAACUUUUUAUUAGUU--GGCACU------GGCACUGGGGCUCACUGGAC--UGUUUGUCCAGCUCGAGCACCAGUUCCAUCUCCAUCACCAUCUCC (--((((((................)))--)))).(------((.(((((.((((.((((((--.....))))))...)))).))))).)))................. ( -35.99) >DroSec_CAF1 59473 97 - 1 U--GCCAGCAAAAAACUUUUUAUUAGUU--GGCACU------GGCACUGGGGCUCACUGGAC--UGUUUGUCCAGCUCCAGCAGCAGUUCCAUCUCCAUCACCAUCACC (--((((((................)))--)))).(------((.((((..(((..((((((--.....))))))....)))..)))).)))................. ( -29.69) >DroSim_CAF1 60030 97 - 1 U--GCCAGCAAAAAACUUUUUAUUAGUU--GGCACU------GGCACUGGGGCUCACUGGAC--UGUUUGUCCAGCUCCAGCACCAGUUCCAUCUCCAUCACCAUCUCC .--((((((................)))--)))(((------((..((((((((....((((--.....))))))))))))..)))))..................... ( -33.49) >DroEre_CAF1 77174 96 - 1 U--GCCAGCAAA-AACUUUUUAUUAGUU--GGCACUGGCACUGGCACUGGGGCUCACUGGAC--UGUUUGUCCAGC------ACCAGCUCCAUCUCCAUCUCCAUCUCC (--((((((...-............)))--)))).(((...(((...(((((((..((((((--.....)))))).------...)))))))...)))...)))..... ( -33.46) >DroYak_CAF1 76339 94 - 1 U--GCCAGCAAA-AACUUUUUAUUAGUU--GGCACUGGCACUGGCACUGGGGCUCACUGGAC--UAUUUGUCCAGC------ACCAACUCCAUCUCCAUUUACAUCC-- (--(((((....-((((.......))))--....)))))).(((...(((((....((((((--.....)))))).------.....)))))...))).........-- ( -28.10) >DroPer_CAF1 69735 99 - 1 UGUGCCAGCAA--AACUUUUUAUUAGUUUCGACGCU------GCUGCCCGGACU--AUGGACUCUGUUUGUCCAUCUGCAACUUCAACUCCAUCUCAAACUCUAGCUUC ...((((((((--((((.......)))))......)------))))..((((..--.(((((.......)))))))))..........................))... ( -15.50) >consensus U__GCCAGCAAA_AACUUUUUAUUAGUU__GGCACU______GGCACUGGGGCUCACUGGAC__UGUUUGUCCAGCUCCAGCACCAGCUCCAUCUCCAUCACCAUCUCC .......((....((((.......))))...)).........((.(((((......((((((.......))))))........))))).)).................. (-14.91 = -15.86 + 0.95)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:14 2006