
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,200,620 – 9,200,713 |
| Length | 93 |
| Max. P | 0.993524 |

| Location | 9,200,620 – 9,200,713 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.05 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -21.62 |
| Energy contribution | -23.62 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
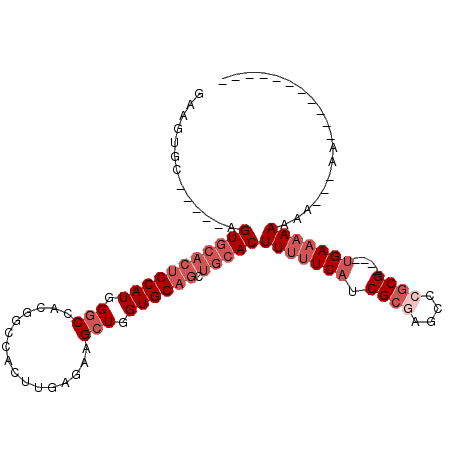
>3L_DroMel_CAF1 9200620 93 + 23771897 GAAGUGC-----AGUGCACUGCAUGGGCCACGGCCACUUGAGAAGCUGGUGCAGCUGCACUUUUUCAUCGCGAGCCCGCG----UGAAAAAAGA---AAAAU--G---AA ...((((-----((((((((((...(((....))).(....)..)).)))))).))))))(((((((.((((....))))----)))))))...---.....--.---.. ( -36.30) >DroPse_CAF1 63784 78 + 1 CAAGUGC-----AGU-----GCAUGGCCCGGAGAUGCUGCUGCUGCUAGUGCUGCUGCACUUUUUCAACGAAGG-CAACG--CAGGACCAA------------------- ..(((((-----(((-----((((.........)))).))))).))).((.(((((((.((((......)))))-))..)--))).))...------------------- ( -23.80) >DroSec_CAF1 54542 87 + 1 GAAGUGC-----AGUGCACUGCAUGGGCCACGGCCACUUGAGAAGCUGGUGCAGCUGCACUUUUUCAUCGCGAGCCCGCG----UGAAAAAAAG---AA----------- ...((((-----((((((((((...(((....))).(....)..)).)))))).))))))(((((((.((((....))))----)))))))...---..----------- ( -36.30) >DroSim_CAF1 55086 87 + 1 GAAGUGC-----AGUGCACUGCAUGGGCCACGGCCACUUGAGAAGCUGGUGCAGCUGCACUUUUUCAUCGCGAGCCCGCG----UGAAAAAAAG---AA----------- ...((((-----((((((((((...(((....))).(....)..)).)))))).))))))(((((((.((((....))))----)))))))...---..----------- ( -36.30) >DroEre_CAF1 72185 96 + 1 GAAGUGC-----AGUGCACUGCAUGGGCCACGGCCACUUGAGAAGCUGGUGCAGCUGCACUUUUUCAUCGCGGGGCCGCG----UGAAAAAAAAUGAAAAAU--G---AA ...((((-----((((((((((...(((....))).(....)..)).)))))).))))))(((((((.((((....))))----)))))))...........--.---.. ( -37.70) >DroYak_CAF1 71149 108 + 1 GAAGUGCAGUGCAGUGCACUGCAUGGGCCACGGCCACUUGAGAUGCUGGUGCAGCUGCACUUUUUCAUCGCAAGUCCGCGUG--UGAAAAAAGAUGAAAAAAGUGAAGAA .....((.((((((((((((((((.(((....))).(....))))).)))))).))))))(((((((((((......))).)--)))))))...........))...... ( -36.90) >consensus GAAGUGC_____AGUGCACUGCAUGGGCCACGGCCACUUGAGAAGCUGGUGCAGCUGCACUUUUUCAUCGCGAGCCCGCG____UGAAAAAAAA___AA___________ .............(((((((((((.(((................))).)))))).)))))(((((((.((((....))))....)))))))................... (-21.62 = -23.62 + 2.00)


| Location | 9,200,620 – 9,200,713 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.05 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -21.08 |
| Energy contribution | -22.45 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9200620 93 - 23771897 UU---C--AUUUU---UCUUUUUUCA----CGCGGGCUCGCGAUGAAAAAGUGCAGCUGCACCAGCUUCUCAAGUGGCCGUGGCCCAUGCAGUGCACU-----GCACUUC ..---.--.....---...(((((((----((((....)))).)))))))((((((.(((((..(((.....)))(((....)))......)))))))-----))))... ( -33.30) >DroPse_CAF1 63784 78 - 1 -------------------UUGGUCCUG--CGUUG-CCUUCGUUGAAAAAGUGCAGCAGCACUAGCAGCAGCAGCAUCUCCGGGCCAUGC-----ACU-----GCACUUG -------------------.((((((.(--.(.((-(....((((....(((((....)))))..))))....))).).).))))))(((-----...-----))).... ( -23.20) >DroSec_CAF1 54542 87 - 1 -----------UU---CUUUUUUUCA----CGCGGGCUCGCGAUGAAAAAGUGCAGCUGCACCAGCUUCUCAAGUGGCCGUGGCCCAUGCAGUGCACU-----GCACUUC -----------..---...(((((((----((((....)))).)))))))((((((.(((((..(((.....)))(((....)))......)))))))-----))))... ( -33.30) >DroSim_CAF1 55086 87 - 1 -----------UU---CUUUUUUUCA----CGCGGGCUCGCGAUGAAAAAGUGCAGCUGCACCAGCUUCUCAAGUGGCCGUGGCCCAUGCAGUGCACU-----GCACUUC -----------..---...(((((((----((((....)))).)))))))((((((.(((((..(((.....)))(((....)))......)))))))-----))))... ( -33.30) >DroEre_CAF1 72185 96 - 1 UU---C--AUUUUUCAUUUUUUUUCA----CGCGGCCCCGCGAUGAAAAAGUGCAGCUGCACCAGCUUCUCAAGUGGCCGUGGCCCAUGCAGUGCACU-----GCACUUC ..---.--...........(((((((----((((....)))).)))))))((((((.(((((..(((.....)))(((....)))......)))))))-----))))... ( -35.00) >DroYak_CAF1 71149 108 - 1 UUCUUCACUUUUUUCAUCUUUUUUCA--CACGCGGACUUGCGAUGAAAAAGUGCAGCUGCACCAGCAUCUCAAGUGGCCGUGGCCCAUGCAGUGCACUGCACUGCACUUC ..........................--..((((((((((.((((.....((((....))))...)))).)))))..))))).....((((((((...)))))))).... ( -34.70) >consensus ___________UU___UUUUUUUUCA____CGCGGGCUCGCGAUGAAAAAGUGCAGCUGCACCAGCUUCUCAAGUGGCCGUGGCCCAUGCAGUGCACU_____GCACUUC ...................(((((((....((((....)))).)))))))(((((.(((((...((((...))))(((....)))..))))))))))............. (-21.08 = -22.45 + 1.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:10 2006