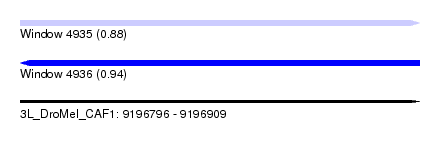
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,196,796 – 9,196,909 |
| Length | 113 |
| Max. P | 0.936083 |

| Location | 9,196,796 – 9,196,909 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -18.21 |
| Energy contribution | -19.30 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9196796 113 + 23771897 UGUCUUGGUUGCGGCAA-GAUUUCUGACAGUUCUGUCAACUAUUUGGGUUGCAACUUGAGCCAGACA--AUUUGCGAAACUAGAACGCCAUCAAAAUACUUGCCACAAUCAAAUAU ....(((((((.(((((-(.....(((((....)))))....(((((.((((((.(((.......))--).))))))..)))))..............)))))).))))))).... ( -32.00) >DroSec_CAF1 50790 112 + 1 UGUCUUGGUUGCGGCAA-AAU-UCUGACAGUUCUGUCAACUAUUUGGGUUGCAACUUGAGCCAGACA--ACUUGCGAAACUAGAACGCCAUCAAAAUACUUGCCGCAAUCAAAUAU ....(((((((((((((-...-..(((((....)))))....(((((.((((((.(((.......))--).))))))..)))))...............))))))))))))).... ( -34.10) >DroSim_CAF1 51307 112 + 1 UGUCUUGGUUGCGGCAA-AAU-UCUGACAGUUCUGUCAACUAUCUGGGUUGCAACUGGAGCCAGACA--ACUUGCGAAACUAGAACGCCAUCAAAAUACUUGCCGCAAUCAAAUAU ....(((((((((((((-...-..(((((....)))))....(((((.(((((((((....)))...--..))))))..)))))...............))))))))))))).... ( -37.20) >DroEre_CAF1 68283 112 + 1 UGUCUUAGUUGCGGCAA-AUU-UCUGACAGUUCUGUCAACUAUUUGGGUUGCAGCUUGAGCCAGACU--ACUUGCGCAACUAGAGCGCCGUCGAAAUACUUGCCACAAUCAAAUAU .......((((.(((((-(((-((.(((.(((((..(((....)))((((((.((..(((.....))--.)..)))))))))))))...))))))))..))))).))))....... ( -29.80) >DroYak_CAF1 67373 114 + 1 UGUCUUGGUUGCGGCAA-AUU-UCUGACAGUUCUGUCAACUAUUUGGGUUGCAACUUGAGCCAGACAGCGCUUGCGAAACUAGAGCGCCAUCAAAAUACUUGCCACAAUCAAAUAU ....(((((((.(((((-...-..(((((....)))))..(((((.((((........)))).((..((((((.........))))))..)).))))).))))).))))))).... ( -33.00) >DroAna_CAF1 52874 91 + 1 GGCUGCCGUUGCGGCAACACU-UCUGACACUUGUGUCACCUAUCUAGGUCUCU------------CUGC-UCUUCGAAUCUGGUU-----------CACUCGGCUCAAUCAACUAU ....((((..((((.......-...((((....))))((((....))))....------------))))-.....((((...)))-----------)...))))............ ( -17.60) >consensus UGUCUUGGUUGCGGCAA_AAU_UCUGACAGUUCUGUCAACUAUUUGGGUUGCAACUUGAGCCAGACA__ACUUGCGAAACUAGAACGCCAUCAAAAUACUUGCCACAAUCAAAUAU ....(((((((.(((((.......(((((....)))))....(((((..(((((.................)))))...)))))...............))))).))))))).... (-18.21 = -19.30 + 1.09)



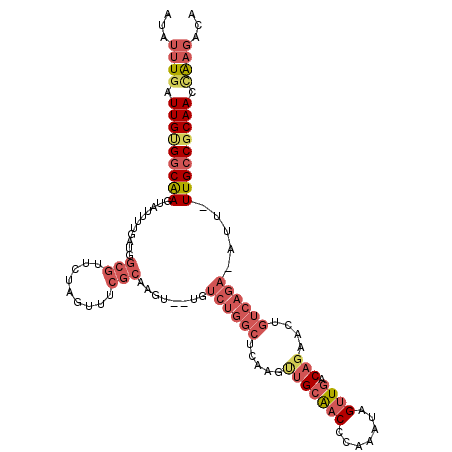
| Location | 9,196,796 – 9,196,909 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.68 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -19.32 |
| Energy contribution | -21.52 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936083 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9196796 113 - 23771897 AUAUUUGAUUGUGGCAAGUAUUUUGAUGGCGUUCUAGUUUCGCAAAU--UGUCUGGCUCAAGUUGCAACCCAAAUAGUUGACAGAACUGUCAGAAAUC-UUGCCGCAACCAAGACA ...((((.((((((((((.(((((..(((((((((.(((..((((.(--((.......))).))))(((.......))))))))))).))))))))))-))))))))).))))... ( -34.10) >DroSec_CAF1 50790 112 - 1 AUAUUUGAUUGCGGCAAGUAUUUUGAUGGCGUUCUAGUUUCGCAAGU--UGUCUGGCUCAAGUUGCAACCCAAAUAGUUGACAGAACUGUCAGA-AUU-UUGCCGCAACCAAGACA ...((((.((((((((((.((((((((...(((((.(((..((((.(--((.......))).))))(((.......))))))))))).))))))-)))-))))))))).))))... ( -36.80) >DroSim_CAF1 51307 112 - 1 AUAUUUGAUUGCGGCAAGUAUUUUGAUGGCGUUCUAGUUUCGCAAGU--UGUCUGGCUCCAGUUGCAACCCAGAUAGUUGACAGAACUGUCAGA-AUU-UUGCCGCAACCAAGACA ...((((.((((((((((.((((((((...(((((.(((.(....)(--(((((((.............))))))))..)))))))).))))))-)))-))))))))).))))... ( -38.12) >DroEre_CAF1 68283 112 - 1 AUAUUUGAUUGUGGCAAGUAUUUCGACGGCGCUCUAGUUGCGCAAGU--AGUCUGGCUCAAGCUGCAACCCAAAUAGUUGACAGAACUGUCAGA-AAU-UUGCCGCAACUAAGACA ...((((.(((((((((((.........((((.......))))....--..((((((.....(((((((.......)))).)))....))))))-.))-))))))))).))))... ( -37.00) >DroYak_CAF1 67373 114 - 1 AUAUUUGAUUGUGGCAAGUAUUUUGAUGGCGCUCUAGUUUCGCAAGCGCUGUCUGGCUCAAGUUGCAACCCAAAUAGUUGACAGAACUGUCAGA-AAU-UUGCCGCAACCAAGACA ...((((.(((((((((((........((((((...........)))))).((((((.....(((((((.......)))).)))....))))))-.))-))))))))).))))... ( -36.90) >DroAna_CAF1 52874 91 - 1 AUAGUUGAUUGAGCCGAGUG-----------AACCAGAUUCGAAGA-GCAG------------AGAGACCUAGAUAGGUGACACAAGUGUCAGA-AGUGUUGCCGCAACGGCAGCC ...(((..((((..(..(..-----------..)..)..))))..)-))..------------....((((....))))((((....))))...-...(((((((...))))))). ( -22.00) >consensus AUAUUUGAUUGUGGCAAGUAUUUUGAUGGCGUUCUAGUUUCGCAAGU__UGUCUGGCUCAAGUUGCAACCCAAAUAGUUGACAGAACUGUCAGA_AUU_UUGCCGCAACCAAGACA ...((((.(((((((((...........(((.........)))........((((((.....(((((((.......)))).)))....)))))).....))))))))).))))... (-19.32 = -21.52 + 2.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:08 2006