
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,195,906 – 9,195,997 |
| Length | 91 |
| Max. P | 0.611749 |

| Location | 9,195,906 – 9,195,997 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.85 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -5.70 |
| Energy contribution | -6.32 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9195906 91 + 23771897 UUAACAAAAG----CCGGCGACAUCAUUGUGCC-----GUGCCAAA----UUGCCAGCAUUUUUUUUU------UAUUUUACCCGACUGGCGGCAAAUUACAUUUAUUUU .........(----(((((.(((....))))))-----........----..((((((..........------..........).))))))))................ ( -19.25) >DroSec_CAF1 49904 92 + 1 UUAACAAAAG----CCGGCGACAUCAUUGUGCC-----GUGCCAAA----UUGCCAGCAGUUUUUUUU---UUUUUUUGUACCCGACUGUCGGCAAAUUACAUUU--UUU .........(----(((((.(((....))))))-----).))....----(((((.((((((......---.............)))))).))))).........--... ( -23.41) >DroSim_CAF1 50414 95 + 1 UUAACAAAAG----CCGGCGACAUCAUUGUGCC-----GUGCCAAA----UUGCCAGCAGUUUUUUUUUUUUUUUUUUGUACCCGACUGUCGGCAAAUUACAUUU--UUU .........(----(((((.(((....))))))-----).))....----(((((.((((((......................)))))).))))).........--... ( -23.25) >DroEre_CAF1 67402 84 + 1 UUAACAAAAG----CCGGCGACAUCAUUGUGUC-----GUGCCAAA----UUGCCAGCAUUUUU----------UUUUGUGCCCGACUG---GCAAAUUACGUUUUUUUU .....(((((----(.((((((((....)))))-----..)))...----((((((((((....----------....)))).....))---)))).....))))))... ( -20.60) >DroYak_CAF1 66472 84 + 1 UUAACAAAAG----CCGGCGACAUCAUUGUGUC-----GUGCCAAA----UGGCCAGCAUUUUU----------UUCUGCUCCCAACUG---GCAAAUUACACCUUUUUU .....(((((----...(((((((....)))))-----))((((..----(((..((((.....----------...)))).)))..))---)).........))))).. ( -21.10) >DroPer_CAF1 57467 103 + 1 UUAACAAAAGGCUACCAGCGAUAUCAUUGUGCCAGAAUGUGCCACAGAAUGUGGCGGAAUAUAUAUUG---GUUAUUCCCCCC----CGGCUGCCAAUUGUUGUAAUUUU .((((((..(((.....(((((...)))))(((.......(((((.....)))))((((((.......---..))))))....----.))).)))..))))))....... ( -28.30) >consensus UUAACAAAAG____CCGGCGACAUCAUUGUGCC_____GUGCCAAA____UUGCCAGCAUUUUUUUUU______UUUUGUACCCGACUG_CGGCAAAUUACAUUU_UUUU ................(((((.....(((.((........))))).....)))))....................................................... ( -5.70 = -6.32 + 0.61)


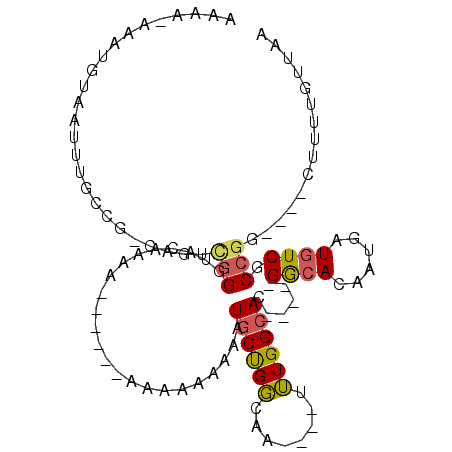
| Location | 9,195,906 – 9,195,997 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.85 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -7.06 |
| Energy contribution | -6.98 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9195906 91 - 23771897 AAAAUAAAUGUAAUUUGCCGCCAGUCGGGUAAAAUA------AAAAAAAAAUGCUGGCAA----UUUGGCAC-----GGCACAAUGAUGUCGCCGG----CUUUUGUUAA ..((((((.((....((((((((((...........------..........))))))..----...))))(-----((((((....))).)))))----).)))))).. ( -22.30) >DroSec_CAF1 49904 92 - 1 AAA--AAAUGUAAUUUGCCGACAGUCGGGUACAAAAAAA---AAAAAAAACUGCUGGCAA----UUUGGCAC-----GGCACAAUGAUGUCGCCGG----CUUUUGUUAA ...--.....(((.((((((.((((..............---.......)))).))))))----.)))((.(-----((((((....))).)))))----)......... ( -21.00) >DroSim_CAF1 50414 95 - 1 AAA--AAAUGUAAUUUGCCGACAGUCGGGUACAAAAAAAAAAAAAAAAAACUGCUGGCAA----UUUGGCAC-----GGCACAAUGAUGUCGCCGG----CUUUUGUUAA ...--...((((.....(((.....))).)))).....................((((((----...(((.(-----((((((....))).)))))----)).)))))). ( -20.90) >DroEre_CAF1 67402 84 - 1 AAAAAAAACGUAAUUUGC---CAGUCGGGCACAAAA----------AAAAAUGCUGGCAA----UUUGGCAC-----GACACAAUGAUGUCGCCGG----CUUUUGUUAA ......(((((((.((((---((((...........----------......))))))))----.)))((.(-----((((......)))))...)----)...)))).. ( -18.03) >DroYak_CAF1 66472 84 - 1 AAAAAAGGUGUAAUUUGC---CAGUUGGGAGCAGAA----------AAAAAUGCUGGCCA----UUUGGCAC-----GACACAAUGAUGUCGCCGG----CUUUUGUUAA ..(((((.((.....(((---(((.(((.((((...----------.....))))..)))----.))))))(-----((((......))))).)).----)))))..... ( -24.20) >DroPer_CAF1 57467 103 - 1 AAAAUUACAACAAUUGGCAGCCG----GGGGGGAAUAAC---CAAUAUAUAUUCCGCCACAUUCUGUGGCACAUUCUGGCACAAUGAUAUCGCUGGUAGCCUUUUGUUAA ........(((((..(((.((((----(((.((((((..---.......))))))(((((.....)))))...))))))).((.((....)).))...)))..))))).. ( -34.50) >consensus AAAA_AAAUGUAAUUUGCCG_CAGUCGGGUACAAAA______AAAAAAAAAUGCUGGCAA____UUUGGCAC_____GGCACAAUGAUGUCGCCGG____CUUUUGUUAA .........................(((.......................((((((........))))))......((((......)))).)))............... ( -7.06 = -6.98 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:06 2006