| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,190,018 – 9,190,114 |
| Length | 96 |
| Max. P | 0.969967 |

| Location | 9,190,018 – 9,190,114 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.78 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9190018 96 - 23771897 -GAGUUGCUGCAAUCUUGGCAAUU-UU-----AUUUGCGCUUUAAGCAA-AUUCCGGCA-AGGCA-AAAGUUGCCACAAAGUUAUUUCAUUAGCUGCAGCC--GU--------G-C---C -.....(((((.....((((((((-((-----((((((.......))))-)).((....-.))..-))))))))))...(((((......)))))))))).--..--------.-.---. ( -28.20) >DroPse_CAF1 50772 104 - 1 ----CGGUUGCAAUCUCGGCAAUU-UU-----AUUUGCGCUUUAAGCAAAAUUCCGGCCAUGGCA-AAAGUUGCCACAAAGUUAUUUCAUUAGCUGCAGCC--AGAUCCUGACACC---C ----.((((((......(((((((-((-----.(((((.......)))))......((....)).-)))))))))....(((((......)))))))))))--.............---. ( -31.70) >DroWil_CAF1 69225 100 - 1 -------------UUUCGGCAAUUUUU-----AUUUGUGCUUUAAGCAA-AUUCCGGCA-AUGCAAAAAGUUGCCACAAAGUUAUUUCAUUAGAUUCAGGCCAGGAUGCUGAUGCUUUUC -------------....((((((((((-----.....((((..((....-.))..))))-.....))))))))))............((((((((((......)))).))))))...... ( -21.80) >DroYak_CAF1 60194 97 - 1 CGAGUAGCUGCAAUCUUGGCAAUU-UU-----AUUUGCGCUUUAAGCAA-AUUCCGGCA-AGGCA-AAAGUUGCCACAAAGUUAUUUCAUUAGCUGCAGCC--GU--------U-C---C .(((..(((((.....((((((((-((-----((((((.......))))-)).((....-.))..-))))))))))...(((((......)))))))))).--.)--------)-)---. ( -32.00) >DroAna_CAF1 46762 107 - 1 -GGAAAGCUGCAAUCUUGGCAAUU-UUAUUUUAUUUGUGCUUUAAGCAA-AUUCCGGCA-AGG-A-AAAGUUGCCACAAAGUUAUUUCAUUAGCUGCAGCC--GGAG-CUGACA-C---C -.....(((((.....((((((((-((.....((((((.......))))-))(((....-.))-)-))))))))))...(((((......)))))))))).--....-......-.---. ( -28.50) >DroPer_CAF1 50567 104 - 1 ----CGGUUGCAAUCUCGGCAAUU-UU-----AUUUGCGCUUUAAGCAAAAUUCCGGCCAUGGCA-AAAGUUGCCACAAAGUUAUUUCAUUAGCUGCAGCC--GGAUCCUGACACC---C ----(((((((......(((((((-((-----.(((((.......)))))......((....)).-)))))))))....(((((......)))))))))))--)............---. ( -33.40) >consensus ____CAGCUGCAAUCUCGGCAAUU_UU_____AUUUGCGCUUUAAGCAA_AUUCCGGCA_AGGCA_AAAGUUGCCACAAAGUUAUUUCAUUAGCUGCAGCC__GGAU_CUGACA_C___C ......(((((......(((((((..........((((.......)))).......((....))....)))))))....(((((......)))))))))).................... (-18.00 = -18.78 + 0.78)

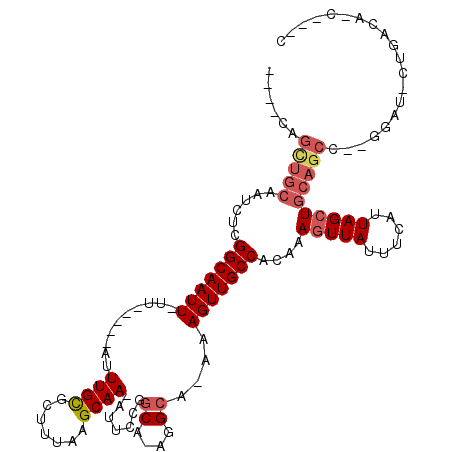

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:00 2006