| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,184,985 – 9,185,118 |
| Length | 133 |
| Max. P | 0.999323 |

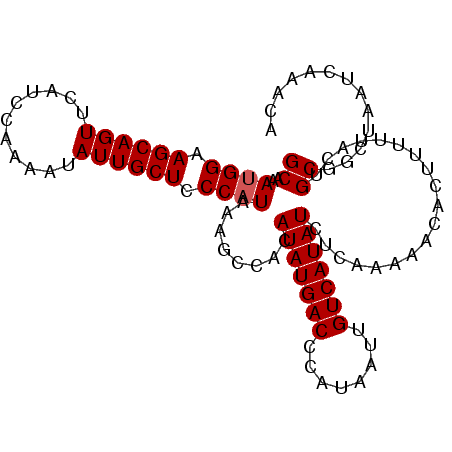
| Location | 9,184,985 – 9,185,086 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9184985 101 - 23771897 GCAAAUGGAAGCAGUUCAUCCAAAAUAUUGCUCCCAUAAAGCCAUAUAUGACCCAUAAUUGUCAUAUCUCAAAAACACUUUUUACUGCGGCUAAUCAAACA ....((((.((((((...........)))))).))))..((((..(((((((........)))))))...((((.....)))).....))))......... ( -19.50) >DroSec_CAF1 39700 101 - 1 GCAAAUGGAAGCAGUUCAUCCAAAAUAUUGCUCCCAUAAAGCGAAAUAUGACCCAUAAUUGUCAUAUCUCAAAAACACUUUUUACUGCGGCUAAUCAAACG (((.((((.((((((...........)))))).)))).....((.(((((((........))))))).))...............)))............. ( -20.80) >DroSim_CAF1 40016 101 - 1 GCAAAUGGAAGCAGUUCAUCCAAAAUAUUGCUCCCAUAAAGCCAUAUAUGACCCAUUAUUGUCAUAUCUCAAAAACACUUUUUACUGCGGCUAUUCAAACG ....((((.((((((...........)))))).))))..((((..(((((((........)))))))...((((.....)))).....))))......... ( -19.50) >DroEre_CAF1 54905 101 - 1 GCAAAUGGAAGCAGUUCAUCCGAAAAAUUGCUCCCCUAAAACCACAUAUGACCCAUAAUAGUCAUAUCUCACAAGCACUUUUUACAGCGGCUAAUCAAACA ((....((.(((((((.........))))))).))..........(((((((........))))))).......))......................... ( -15.20) >DroYak_CAF1 55040 101 - 1 GCAAAUGGAAGCAGUUCAUCCAAAAUAUUGCUUCCAUAAAAGCACAUAUGACCCAUAAUUGUCAUAUCUCAAAAACACUUUUUACAGCGACUGAUCAAACA ((..(((((((((((...........)))))))))))........(((((((........)))))))...................))............. ( -21.80) >consensus GCAAAUGGAAGCAGUUCAUCCAAAAUAUUGCUCCCAUAAAGCCACAUAUGACCCAUAAUUGUCAUAUCUCAAAAACACUUUUUACUGCGGCUAAUCAAACA ((..((((.((((((...........)))))).))))........(((((((........)))))))...................))............. (-16.34 = -16.54 + 0.20)

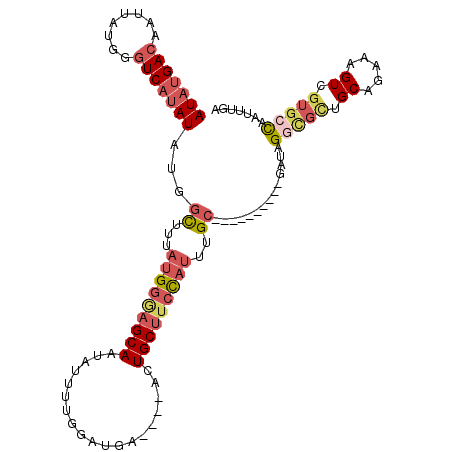
| Location | 9,185,019 – 9,185,118 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -17.10 |
| Energy contribution | -18.69 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9185019 99 + 23771897 AUAUGACAAUUAUGGGUCAUAUAUGGCUUUAUGGGAGCAAUAUUUUGGAUGA----ACUGCUUCCAUUUGC----------GAUUGCCGCUGCAGAAAGUCGUGCCAAUAUGA (((((((........))))))).((((...(((((((((.............----..)))))))))..((----------((((..(......)..))))))))))...... ( -28.16) >DroSec_CAF1 39734 99 + 1 AUAUGACAAUUAUGGGUCAUAUUUCGCUUUAUGGGAGCAAUAUUUUGGAUGA----ACUGCUUCCAUUUGC----------GAUAGGCGCUGCAGAAAGUCGUGCCAAUUUGA (((((((........))))))).((((...(((((((((.............----..)))))))))..))----------))..(((((.((.....)).)))))....... ( -32.76) >DroSim_CAF1 40050 99 + 1 AUAUGACAAUAAUGGGUCAUAUAUGGCUUUAUGGGAGCAAUAUUUUGGAUGA----ACUGCUUCCAUUUGC----------GAUAGGUGCUGCAGAAAGUCGUGCCAAUUUGA .((((((........))))))(((.((...(((((((((.............----..)))))))))..))----------.)))((..(.((.....)).)..))....... ( -28.96) >DroEre_CAF1 54939 99 + 1 AUAUGACUAUUAUGGGUCAUAUGUGGUUUUAGGGGAGCAAUUUUUCGGAUGA----ACUGCUUCCAUUUGC----------GAUAGGCGGUGCAGAAAGUCGUGCCAAAUUGA .(((((((......)))))))(((.((.....(((((((.(((.......))----).)))))))....))----------.)))...((..(........)..))....... ( -26.70) >DroYak_CAF1 55074 99 + 1 AUAUGACAAUUAUGGGUCAUAUGUGCUUUUAUGGAAGCAAUAUUUUGGAUGA----ACUGCUUCCAUUUGC----------GAUAGGCGUUGCAGAAAGUCGAGCUAAAUUGA (((((((........)))))))..((((...((((((((.............----..))))))))(((((----------(((....)))))))).....))))........ ( -26.56) >DroAna_CAF1 41568 100 + 1 --AUGAUAAC-----CUC------AGCUGUUUGUUUGCAAUGAGCUGGAUGAGUUUAAUGCUUUUACUUGCUUCAUAUUCAUAUAGAAGCUGCAGAAAGUCGUGCCAAUUUGA --........-----.((------((((..(((....)))..))))))..(((((..((((((((....(((((.(((....))))))))....))))).)))...))))).. ( -21.20) >consensus AUAUGACAAUUAUGGGUCAUAUAUGGCUUUAUGGGAGCAAUAUUUUGGAUGA____ACUGCUUCCAUUUGC__________GAUAGGCGCUGCAGAAAGUCGUGCCAAUUUGA (((((((........)))))))...((...(((((((((...................)))))))))..))..............(((((.((.....)).)))))....... (-17.10 = -18.69 + 1.59)

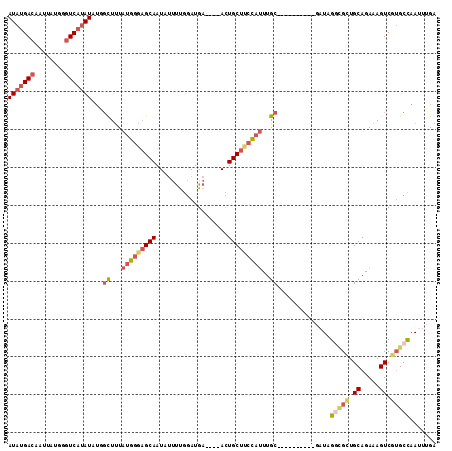
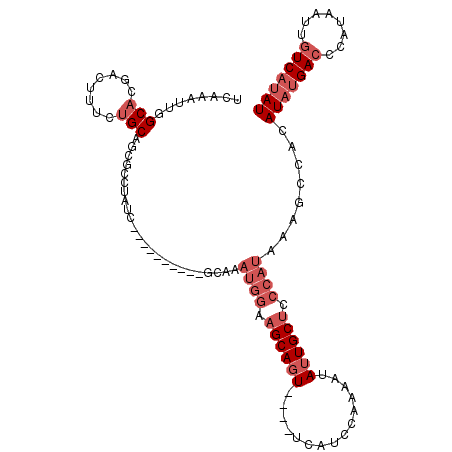
| Location | 9,185,019 – 9,185,118 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -9.46 |
| Energy contribution | -11.21 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9185019 99 - 23771897 UCAUAUUGGCACGACUUUCUGCAGCGGCAAUC----------GCAAAUGGAAGCAGU----UCAUCCAAAAUAUUGCUCCCAUAAAGCCAUAUAUGACCCAUAAUUGUCAUAU ......((((.............(((.....)----------))..((((.((((((----...........)))))).))))...)))).(((((((........))))))) ( -24.50) >DroSec_CAF1 39734 99 - 1 UCAAAUUGGCACGACUUUCUGCAGCGCCUAUC----------GCAAAUGGAAGCAGU----UCAUCCAAAAUAUUGCUCCCAUAAAGCGAAAUAUGACCCAUAAUUGUCAUAU .......(((.(..(.....)..).)))..((----------((..((((.((((((----...........)))))).))))...)))).(((((((........))))))) ( -25.30) >DroSim_CAF1 40050 99 - 1 UCAAAUUGGCACGACUUUCUGCAGCACCUAUC----------GCAAAUGGAAGCAGU----UCAUCCAAAAUAUUGCUCCCAUAAAGCCAUAUAUGACCCAUUAUUGUCAUAU ......((((.............((.......----------))..((((.((((((----...........)))))).))))...)))).(((((((........))))))) ( -22.10) >DroEre_CAF1 54939 99 - 1 UCAAUUUGGCACGACUUUCUGCACCGCCUAUC----------GCAAAUGGAAGCAGU----UCAUCCGAAAAAUUGCUCCCCUAAAACCACAUAUGACCCAUAAUAGUCAUAU ......(((..........(((..........----------)))...((.((((((----(.........))))))).))......))).(((((((........))))))) ( -15.50) >DroYak_CAF1 55074 99 - 1 UCAAUUUAGCUCGACUUUCUGCAACGCCUAUC----------GCAAAUGGAAGCAGU----UCAUCCAAAAUAUUGCUUCCAUAAAAGCACAUAUGACCCAUAAUUGUCAUAU ........(((........(((..........----------))).(((((((((((----...........)))))))))))...)))..(((((((........))))))) ( -22.90) >DroAna_CAF1 41568 100 - 1 UCAAAUUGGCACGACUUUCUGCAGCUUCUAUAUGAAUAUGAAGCAAGUAAAAGCAUUAAACUCAUCCAGCUCAUUGCAAACAAACAGCU------GAG-----GUUAUCAU-- ............((((((..((.((((((((....))).)))))..((((.(((..............)))..)))).........)).------)))-----))).....-- ( -15.74) >consensus UCAAAUUGGCACGACUUUCUGCAGCGCCUAUC__________GCAAAUGGAAGCAGU____UCAUCCAAAAUAUUGCUCCCAUAAAGCCACAUAUGACCCAUAAUUGUCAUAU ........(((........)))........................((((.((((((...............)))))).))))........(((((((........))))))) ( -9.46 = -11.21 + 1.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:59 2006