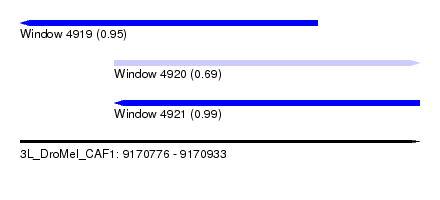
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,170,776 – 9,170,933 |
| Length | 157 |
| Max. P | 0.992016 |

| Location | 9,170,776 – 9,170,893 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -12.34 |
| Energy contribution | -11.46 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9170776 117 - 23771897 AACAUAUUGUCUUUUCGAGCUUAUUUUAUCCUCGGAGCUUUACCAAAAUAUCCAUCUUUUAUCCAAAGCUCCAAGCUUUGCAUUGC---AAGCUUUUUCAAAGUCUUCGGUUGCGUAAAA ......(((..(..((((((((...........((((((((...((((........))))....))))))))((((((.((...))---)))))).....))))..))))..)..))).. ( -20.30) >DroSec_CAF1 25701 110 - 1 AACAGAUUGUCCUUUCAAGCUUAUUUUAUCCUUGGAGCUUUACCACAAUUUCCACCUUUUAUCCAAAGCUCCAAGCUUUGCUUCGC---AAGCUUUUUCAUAGUCUUCGGUC-------A ...(((((((......((((((........(((((((((((.......................)))))))))))....((...))---))))))....)))))))......-------. ( -22.40) >DroSim_CAF1 26032 110 - 1 AACAGAUUGUCCUUUCAAGCUUAUUUUAUCCUUGGAGCUUUACCACAGUUUCUAUCUCUUAUCCAAAGCUCCAAGCUUUGCACUGC---AAGCUUUUUCAAAGUCUUCGGUC-------G ....(((((..((((.((((((........(((((((((((.....((.........)).....)))))))))))....((...))---))))))....))))....)))))-------. ( -23.40) >DroEre_CAF1 38549 112 - 1 AACAAAUUGUCCUUUCAAGUUUACAUUAUCCUUGGAGCUUUA--------UCCAGCUUUUAUCCGAAGCUCUUAGCAUUACUAGGAAAAAAGCUGUUCCAGAGUCUUUGGUUGUGUAAAA ...................(((((((...((..(((.((((.--------..((((((((.(((.((((.....)).))....))).))))))))....)))))))..))..))))))). ( -27.50) >DroYak_CAF1 40144 111 - 1 AACAAAUUGUCCUUUCAAGUUCAUAUUAUCCUUGGAGCUUUA--------UGCAGCUUUUAUCCAAAGCUCUUAGCUUUACUAAGCAGAAAGCUUUUUCAGAGUCUUUGGUUGUGUAA-A .((((...........(((((((((...((....))....))--------)).)))))....((((((((((.((((((.((....)))))))).....)))).))))))))))....-. ( -24.40) >consensus AACAAAUUGUCCUUUCAAGCUUAUUUUAUCCUUGGAGCUUUACCA_A_U_UCCAGCUUUUAUCCAAAGCUCCAAGCUUUGCUAUGC___AAGCUUUUUCAAAGUCUUCGGUUG_GUAA_A ..........((....((((((...........((((((((.......................))))))))((((((...........)))))).....))).))).)).......... (-12.34 = -11.46 + -0.88)



| Location | 9,170,813 – 9,170,933 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9170813 120 + 23771897 CAAAGCUUGGAGCUUUGGAUAAAAGAUGGAUAUUUUGGUAAAGCUCCGAGGAUAAAAUAAGCUCGAAAAGACAAUAUGUUGUGAAAUUCUCUAUUUACUUCAUUGAAGCUUUAAAUAUUU .....((((((((((((..(((((........))))).))))))))))))((((....((((((((...(((.....)))(((((........)))))....))).)))))....)))). ( -27.30) >DroSec_CAF1 25731 120 + 1 CAAAGCUUGGAGCUUUGGAUAAAAGGUGGAAAUUGUGGUAAAGCUCCAAGGAUAAAAUAAGCUUGAAAGGACAAUCUGUUGUGAAAUUCUCUAUUUAAGUCUUUAAAGCUUUAAACAUUU .....((((((((((((..((...(((....))).)).))))))))))))........((((((..((((((.....((.(.(.....).).))....)))))).))))))......... ( -27.80) >DroSim_CAF1 26062 120 + 1 CAAAGCUUGGAGCUUUGGAUAAGAGAUAGAAACUGUGGUAAAGCUCCAAGGAUAAAAUAAGCUUGAAAGGACAAUCUGUUGUGAAAUUCUCUAUUUACUUCUUUGAAGCUUUAAAAAUGU .....((((((((((((....(.((.......)).)..))))))))))))........((((((.(((((((.....)))(((((........)))))..)))).))))))......... ( -29.00) >DroEre_CAF1 38589 111 + 1 UAAUGCUAAGAGCUUCGGAUAAAAGCUGGA--------UAAAGCUCCAAGGAUAAUGUAAACUUGAAAGGACAAUUUGUUGUGAAAUUGCCUAUUUAUUU-UUUGAAGCUUUACAAGUUU ........(((((((((((....((((...--------...)))).((((...........))))..(((.((((((......)))))))))........-)))))))))))........ ( -24.30) >DroYak_CAF1 40183 109 + 1 UAAAGCUAAGAGCUUUGGAUAAAAGCUGCA--------UAAAGCUCCAAGGAUAAUAUGAACUUGAAAGGACAAUUUGUUGUGAAAUUGUCUAUUUAAUA-UU--GAGCUUUGCAAGGGU ....(((...((((((.....))))))(((--------..((((((((((...........))))...(((((((((......)))))))))........-..--)))))))))...))) ( -27.40) >consensus CAAAGCUUGGAGCUUUGGAUAAAAGAUGGA_A_U_UGGUAAAGCUCCAAGGAUAAAAUAAGCUUGAAAGGACAAUCUGUUGUGAAAUUCUCUAUUUAAUUCUUUGAAGCUUUAAAAAUUU ((((((.....)))))).....................(((((((.((((...........)))).....((((....))))........................)))))))....... (-14.42 = -14.10 + -0.32)


| Location | 9,170,813 – 9,170,933 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.64 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9170813 120 - 23771897 AAAUAUUUAAAGCUUCAAUGAAGUAAAUAGAGAAUUUCACAACAUAUUGUCUUUUCGAGCUUAUUUUAUCCUCGGAGCUUUACCAAAAUAUCCAUCUUUUAUCCAAAGCUCCAAGCUUUG .......((((((((..(((((((((...(((((....((((....)))).)))))....)))))))))....((((((((...((((........))))....)))))))))))))))) ( -23.00) >DroSec_CAF1 25731 120 - 1 AAAUGUUUAAAGCUUUAAAGACUUAAAUAGAGAAUUUCACAACAGAUUGUCCUUUCAAGCUUAUUUUAUCCUUGGAGCUUUACCACAAUUUCCACCUUUUAUCCAAAGCUCCAAGCUUUG .........((((((.((((.........((.(((((......))))).)))))).))))))........(((((((((((.......................)))))))))))..... ( -22.50) >DroSim_CAF1 26062 120 - 1 ACAUUUUUAAAGCUUCAAAGAAGUAAAUAGAGAAUUUCACAACAGAUUGUCCUUUCAAGCUUAUUUUAUCCUUGGAGCUUUACCACAGUUUCUAUCUCUUAUCCAAAGCUCCAAGCUUUG .........((((((.((((.........((.(((((......))))).)))))).))))))........(((((((((((.....((.........)).....)))))))))))..... ( -22.90) >DroEre_CAF1 38589 111 - 1 AAACUUGUAAAGCUUCAAA-AAAUAAAUAGGCAAUUUCACAACAAAUUGUCCUUUCAAGUUUACAUUAUCCUUGGAGCUUUA--------UCCAGCUUUUAUCCGAAGCUCUUAGCAUUA ......((((((((((((.-.........((((((((......)))))))).......(....).......)))))))))))--------)..((((((.....)))))).......... ( -21.50) >DroYak_CAF1 40183 109 - 1 ACCCUUGCAAAGCUC--AA-UAUUAAAUAGACAAUUUCACAACAAAUUGUCCUUUCAAGUUCAUAUUAUCCUUGGAGCUUUA--------UGCAGCUUUUAUCCAAAGCUCUUAGCUUUA ....(((((((((((--..-.........((((((((......))))))))....((((...........))))))))))..--------))))).........(((((.....))))). ( -23.50) >consensus AAAUUUUUAAAGCUUCAAAGAAGUAAAUAGAGAAUUUCACAACAAAUUGUCCUUUCAAGCUUAUUUUAUCCUUGGAGCUUUACCA_A_U_UCCAGCUUUUAUCCAAAGCUCCAAGCUUUG .......((((((((..............((.(((((......))))).))......................((((((((.......................)))))))))))))))) (-14.48 = -14.64 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:54 2006