
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,167,521 – 9,167,648 |
| Length | 127 |
| Max. P | 0.852594 |

| Location | 9,167,521 – 9,167,628 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -13.33 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.45 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536726 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9167521 107 - 23771897 AUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCACUUUUCCAGCACCUCUCCCAUUCC .....................((((....................((((((((....))))))))...((......)).............))))............ ( -13.00) >DroPse_CAF1 25160 96 - 1 AUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGGCUCCACUCCUCAAUCCCC----------- .......................((((((((((....))))...(((((((((....))))))))).......)))))).................----------- ( -15.90) >DroEre_CAF1 35192 103 - 1 AUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCACUUUUCCAGGCCC----CAUCUCC ...................(((.(((...................((((((((....))))))))...((......)).............)))..----))).... ( -13.00) >DroYak_CAF1 36771 95 - 1 AUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCACCCCU--------CCC----UCCCUCC .................((((((..................))))))((((((....)))))).....((......)).......--------...----....... ( -10.37) >DroAna_CAF1 24368 86 - 1 AUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUCCCCCCAUAC--------------------- ....................((((.(...((((....))))....((((((((....))))))))).))))...............--------------------- ( -11.80) >DroPer_CAF1 25135 96 - 1 AUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGGCUCCACUCCUCAAUCCCC----------- .......................((((((((((....))))...(((((((((....))))))))).......)))))).................----------- ( -15.90) >consensus AUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGCCACCACUC_UC_A_CCCC___________ ....................((((.(...((((....))))....((((((((....))))))))).)))).................................... (-10.70 = -10.45 + -0.25)

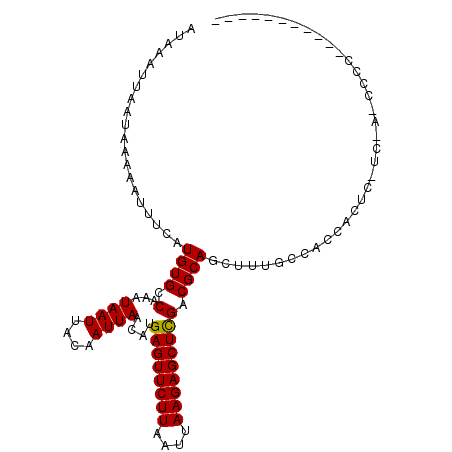

| Location | 9,167,548 – 9,167,648 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -21.75 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9167548 100 - 23771897 UUGCC---GG------------CGCUGCCAGCGGUAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCA .....---((------------((((((....((((((((((......))))...))))))...................((((((((....))))))))...)))))...))). ( -25.20) >DroPse_CAF1 25176 109 - 1 UUGCU---GUUGGUUUUG---CUGUUGCGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGGCU .((((---((((((((((---(....))))..((((((((((......))))...))))))..........))))))))(((((((((....)))))))))..)))((....)). ( -28.30) >DroEre_CAF1 35215 100 - 1 UUGCC---AG------------CGCUGGCAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUUGACGCAGCUUUGCCA .((((---(.------------...)))))((((((((((((......))))...))))))...................((((((((....))))))))...)).......... ( -23.90) >DroWil_CAF1 31814 115 - 1 CUGCUGCUUCUGGUUCUGCUGCUGUUGUGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCACGACUCAGCUUUUGCU ..((((..((..(((((((((((......)))))))....(((((......(((((((..................)))))))......)))))))))..))..))))....... ( -26.77) >DroAna_CAF1 24374 100 - 1 UUGCC---UU------------CGUUGCGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUCCCC .....---..------------.((((((...((((((((((......))))...))))))..................(((((((((....))))))))).))))))....... ( -26.40) >DroPer_CAF1 25151 109 - 1 UUGCU---GUUGGUUUUG---CUGUUGCGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGGCU .((((---((((((((((---(....))))..((((((((((......))))...))))))..........))))))))(((((((((....)))))))))..)))((....)). ( -28.30) >consensus UUGCC___GU____________CGUUGCGAGCGGCAUAAAUUAAUAAAAAUUUCAUGUGCCAAAUAAUUACAAUUAACAUGAGUUCUUAAUUAAGAGCUCGACGCAGCUUUGCCU .......................((((((...((((((((((......))))...))))))..................(((((((((....))))))))).))))))....... (-21.75 = -22.00 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:51 2006