
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,165,848 – 9,165,952 |
| Length | 104 |
| Max. P | 0.999326 |

| Location | 9,165,848 – 9,165,952 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.17 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
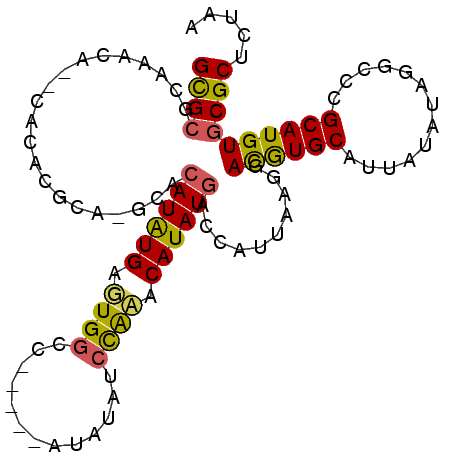
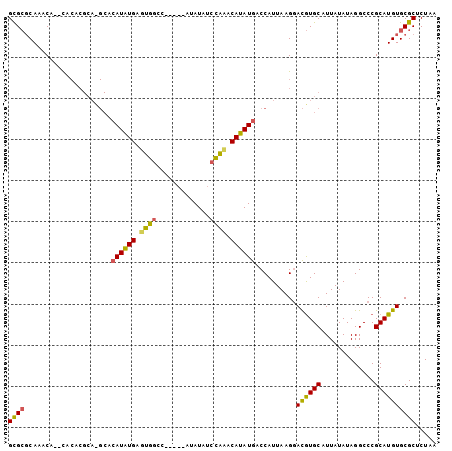
>3L_DroMel_CAF1 9165848 104 + 23771897 GCGCUCAAACACACACACGCAUGCACAUGUGAGUGGCCGCCAUACAUAUCCACACAUAUGACCAUUAAGGACGUGCAUUAUAUAGGCCCGCAUGUGCGCUCUAA ((((..............))..(((((((((...((((......(((((......))))).((.....))..............)))))))))))))))..... ( -27.34) >DroSec_CAF1 20864 97 + 1 GCGCGCACACA--CACACGCACGCACAUAUGAGUGGCC-----AUAUAUCCAAACAUAUGCCCAUUAAGAACGUGCAUUAUAUAGGCCCGCAUGUGCGCUCUAA ..(((((((..--.....(((((........(((((.(-----((((........))))).))))).....))))).........(....).)))))))..... ( -26.82) >DroAna_CAF1 22560 83 + 1 GUGGGCAGACA--U------------AUAUGUAUGAGG-----AUAUA--UGUACAUAUAUCCAUUAAGGAUAUGCAUUAUAUAGGCCCGCAUGUGCGCUCUUA ((((((..(((--(------------((((.(....).-----)))))--)))...(((((((.....)))))))..........))))))............. ( -25.20) >consensus GCGCGCAAACA__CACACGCA_GCACAUAUGAGUGGCC_____AUAUAUCCAAACAUAUGACCAUUAAGGACGUGCAUUAUAUAGGCCCGCAUGUGCGCUCUAA ((((.....................((((((.((((.............)))).))))))..........((((((.............))))))))))..... (-15.38 = -15.17 + -0.21)

| Location | 9,165,848 – 9,165,952 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.15 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -14.19 |
| Energy contribution | -16.70 |
| Covariance contribution | 2.51 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
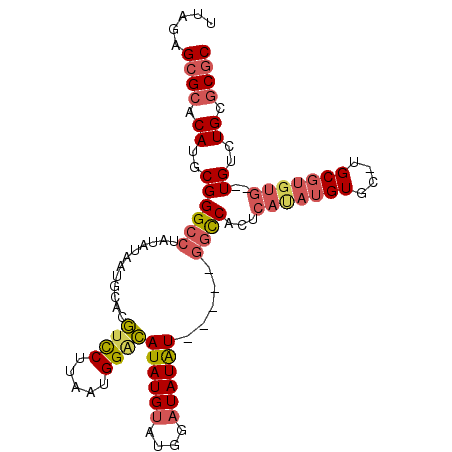
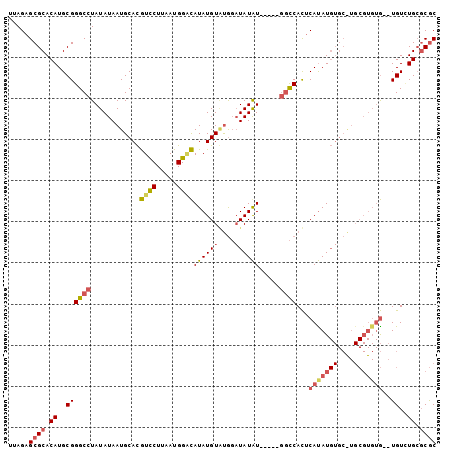
>3L_DroMel_CAF1 9165848 104 - 23771897 UUAGAGCGCACAUGCGGGCCUAUAUAAUGCACGUCCUUAAUGGUCAUAUGUGUGGAUAUGUAUGGCGGCCACUCACAUGUGCAUGCGUGUGUGUGUUUGAGCGC ...(((((((((((((.(((((((((.(((((((((.....))....)))))))..)))))).))).((.((......))))...)))))))))))))...... ( -35.50) >DroSec_CAF1 20864 97 - 1 UUAGAGCGCACAUGCGGGCCUAUAUAAUGCACGUUCUUAAUGGGCAUAUGUUUGGAUAUAU-----GGCCACUCAUAUGUGCGUGCGUGUG--UGUGUGCGCGC .....(((((((((((.((.......((((((((......(((.(((((((....))))))-----).))).....))))))))..)).))--))))))))).. ( -40.80) >DroAna_CAF1 22560 83 - 1 UAAGAGCGCACAUGCGGGCCUAUAUAAUGCAUAUCCUUAAUGGAUAUAUGUACA--UAUAU-----CCUCAUACAUAU------------A--UGUCUGCCCAC .......((....))((((.......((..((((((.....))))))..))(((--(((((-----........))))------------)--)))..)))).. ( -20.60) >consensus UUAGAGCGCACAUGCGGGCCUAUAUAAUGCACGUCCUUAAUGGACAUAUGUAUGGAUAUAU_____GGCCACUCAUAUGUGC_UGCGUGUG__UGUCUGCGCGC .....((((.((..((((((............((((.....))))((((((....)))))).....))))...(((((((....)))))))..))..)).)))) (-14.19 = -16.70 + 2.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:50 2006