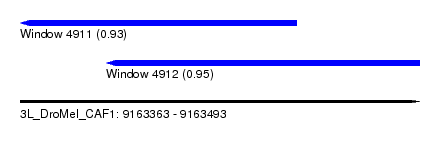
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,163,363 – 9,163,493 |
| Length | 130 |
| Max. P | 0.952353 |

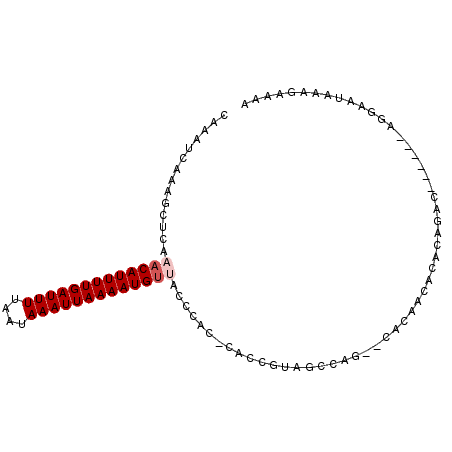
| Location | 9,163,363 – 9,163,453 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -13.69 |
| Consensus MFE | -8.40 |
| Energy contribution | -8.90 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9163363 90 - 23771897 CAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCAC-CACCGUAGCCAG--CACAACACACAGAC------AGGAAUAAAGAAAA .........(((..(((((((((((((....)))))))))))))....((-....))....))--).............------.............. ( -10.60) >DroSec_CAF1 18475 90 - 1 CAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCAC-CACCGUAGCCAG--CACAACACACAGAC------AGGAAUAAAGAAAA .........(((..(((((((((((((....)))))))))))))....((-....))....))--).............------.............. ( -10.60) >DroSim_CAF1 18648 90 - 1 CAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUAACCCAC-CACCAUAGCCAG--CACAACACACAGAC------AGGAAUAAAGAAAA ...............((((((((((((....)))))))))))).......-............--..............------.............. ( -9.00) >DroEre_CAF1 30936 75 - 1 CAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCC---AC-------------CAA--CGUAGAC------GGGUACAAGGAAAA ....((........(((((((((((((....)))))))))))))((((.---..-------------...--.......------)))).....))... ( -13.32) >DroYak_CAF1 32473 89 - 1 CAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGCCACCACC-CAC---AGCGAACACACAACCCACAGAC------AGGAAUAAAGAAAA ....((..........(((((((((((....)))))))))))........-...---..............((......------.))......))... ( -7.80) >DroAna_CAF1 19538 97 - 1 CAAAUCGGAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCGGCCCACGCCCCCCU--CCCAACUUGGAGAUGGGGCCGGAAAAAAGGAAAA ....((((.(((..(((((((((((((....)))))))))))))....)))))..(((((.((--((......))))..)))))..))........... ( -30.80) >consensus CAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCAC_CACCGUAGCCAG__CACAACACACAGAC______AGGAAUAAAGAAAA ..............(((((((((((((....)))))))))))))....................................................... ( -8.40 = -8.90 + 0.50)

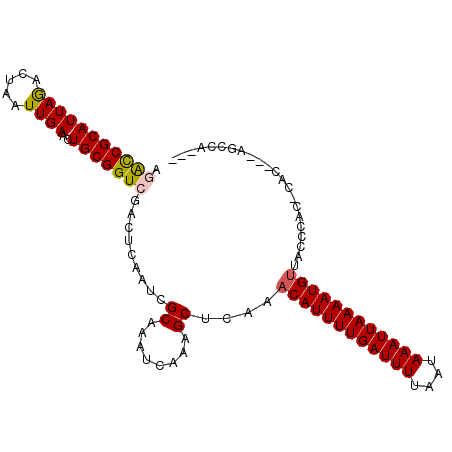
| Location | 9,163,391 – 9,163,493 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -21.71 |
| Consensus MFE | -17.67 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9163391 102 - 23771897 AGACCGCAUUAGACUAAUUGAACUGCGGUCGACUCAAUCGCAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCAC-CACCGUAGCCAG-- .(((((((...............))))))).........((........))...(((((((((((((....)))))))))))))......-............-- ( -21.76) >DroSim_CAF1 18676 102 - 1 AGACCGCAUUAGACUAAUUGAACUGCGGUCGACUCAAUCGCAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUAACCCAC-CACCAUAGCCAG-- .(((((((...............))))))).........((........))....((((((((((((....)))))))))))).......-............-- ( -20.86) >DroEre_CAF1 30960 91 - 1 AAACCGCAUUAGGCUAAUUGAACUGCGGUCGACUCAAUCGCAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCC---AC----------- ...........((((..((((..((((((.......))))))..))))))))..(((((((((((((....))))))))))))).....---..----------- ( -21.50) >DroWil_CAF1 25336 92 - 1 -CGUCGCAUUAAACUAAUUGAAAUGCGGUUCAACAAAUUGCAAAUCAAAGCUCGAACAUUUUGAUUUUAAUAAAUUAAAAUGUGGCCAAC-AAG----------- -.(((............((((..(((((((.....)))))))..)))).......((((((((((((....)))))))))))))))....-...----------- ( -18.70) >DroYak_CAF1 32501 101 - 1 AGACCGCAUUAGACUAAUUGAACUGCGGUCGACUCAAUCGCAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGCCACCACC-CAC---AGCGAACA .(((((((...............))))))).......((((...............(((((((((((....)))))))))))........-...---.))))... ( -21.09) >DroAna_CAF1 19572 103 - 1 AGACCGCAUUAGACUAAUUGAACUGCGGUCGACUCAAUCGCAAAUCGGAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCGGCCCACGCCCCCCU-- .(((((((...............)))))))................((.(((..(((((((((((((....)))))))))))))....)))))..........-- ( -26.36) >consensus AGACCGCAUUAGACUAAUUGAACUGCGGUCGACUCAAUCGCAAAUCAAAGCUCAAACAUUUUGAUUUUAAUAAAUUAAAAUGUUACCCAC_CAC___AGCCA___ .(((((((((((.....))))..))))))).........((........))....((((((((((((....))))))))))))...................... (-17.67 = -17.75 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:46 2006