
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,153,245 – 9,153,373 |
| Length | 128 |
| Max. P | 0.663046 |

| Location | 9,153,245 – 9,153,346 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.64 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -23.11 |
| Energy contribution | -24.03 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
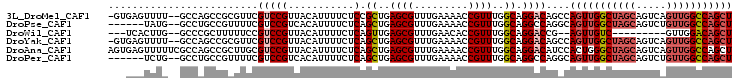
>3L_DroMel_CAF1 9153245 101 + 23771897 -GUGAGUUUU--GCCAGCCGCGUUCGUCCGUUACAUUUUCUCCGCUGAGCGUUUGAAAACCGUUUGGCAGGACAGCCAGUUGGCUAGCAGUCAGUUGGCCAGCU -....(((((--(((((.((..((((..((((.((..........))))))..))))...)).))))))))))....(((((((((((.....))))))))))) ( -38.20) >DroPse_CAF1 9112 96 + 1 ------UAUG--GCCUGCCGUUUUCGUCCGUCACAUUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGCCAGGCAGUUGGCUAGCAGUCUGUUGGCCAGCU ------..((--((((((((((((((..(((((............))).))..))))))).....)))))))))...(((((((((((.....))))))))))) ( -43.50) >DroWil_CAF1 2600 88 + 1 ---UCACUUG--GCCCGCUUUUUCCGUCCGUUACAUUUUCUCAGUUGAGCGUUUGAACACCGUUUGGCAGGACCG--AGUUGUC---------GUUGGACAGCU ---....(((--(((.(((.....((...(((.((..(.(((....))).)..)))))..))...))).)).)))--)((((((---------....)))))). ( -21.20) >DroYak_CAF1 21839 101 + 1 -GUGAGUUUU--GCCAGCCGCGUUCGUCCGUUACAUUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGACAGCCAGUUGGCUAGCAGUCAGUUGGCCAGCU -....(((((--(((((.((..((((..((((.((..........))))))..))))...)).))))))))))....(((((((((((.....))))))))))) ( -38.20) >DroAna_CAF1 8862 104 + 1 AGUGAGUUUUUCGCCAGCCGCUUGCGUCCGUUACAUUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGACAUCCACUGGGCUAGCAGUCAGUUGGCCAGCU ............((((((.(.((((((((..............((..((((.........))))..)).((....))...))))..)))).).))))))..... ( -29.80) >DroPer_CAF1 9116 96 + 1 ------UCUG--GCCUGCCGUUUUCGUCCGUCACAUUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGCCAGGCAGUUGGCUAGCAGUCUGUUGGCCAGCU ------((((--((((((((((((((..(((((............))).))..))))))).....))))))))))).(((((((((((.....))))))))))) ( -46.50) >consensus ___GAGUUUG__GCCAGCCGCUUUCGUCCGUUACAUUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGACAGCCAGUUGGCUAGCAGUCAGUUGGCCAGCU .........................(((((...........).((..((((.........))))..)).))))....(((((((((((.....))))))))))) (-23.11 = -24.03 + 0.92)

| Location | 9,153,277 – 9,153,373 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.02 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -26.92 |
| Energy contribution | -27.58 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9153277 96 + 23771897 UUUUCUCCGCUGAGCGUUUGAAAACCGUUUGGCAGGACAGCCAGUUGGCUAGCAGUCAGUUGGCCAGCUAGGAGCCG------AG-----CAGUCCUUGCAGGACGA (((((..(((...)))...)))))..(((((((.((....))(((((((((((.....)))))))))))....))))------))-----).((((.....)))).. ( -38.80) >DroPse_CAF1 9139 107 + 1 UUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGCCAGGCAGUUGGCUAGCAGUCUGUUGGCCAGCUAGGAGCCAAAGCCGAGCGUGCCAGGCUUUGCAGACACA ....(((....))).(((((......((((((((.((..((((((((((((((.....))))))))))).(....)...)))..)).))))))))....)))))... ( -45.80) >DroEre_CAF1 20618 96 + 1 UUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGACAGCCAGUUGGCUAGCAGUCAGUUGGCCAGCUAGGAGCCG------AG-----CAGGCCUUGCAGGACGA (((((...((...))....))))).((((..(((((....(((((((((((((.....))))))))))).)).(((.------..-----..))))))))..)))). ( -37.50) >DroYak_CAF1 21871 96 + 1 UUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGACAGCCAGUUGGCUAGCAGUCAGUUGGCCAGCUAGGAGCCG------AG-----CAGGCCUUGCAGGACGA (((((...((...))....))))).((((..(((((....(((((((((((((.....))))))))))).)).(((.------..-----..))))))))..)))). ( -37.50) >DroAna_CAF1 8897 85 + 1 UUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGACAUCCACUGGGCUAGCAGUCAGUUGGCCAGCUAGGAGCCG------AG-----CCGAGC----------- (((((...((...))....)))))..(((((((.((...(((....(((((((.....))))))).....))).)).------.)-----))))))----------- ( -28.50) >DroPer_CAF1 9143 107 + 1 UUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGCCAGGCAGUUGGCUAGCAGUCUGUUGGCCAGCUAGGAGCCAAAGCCGAGCGUGCCAGGCUUUGCAGACACA ....(((....))).(((((......((((((((.((..((((((((((((((.....))))))))))).(....)...)))..)).))))))))....)))))... ( -45.80) >consensus UUUUCUCAGCUGAGCGUUUGAAAACCGUUUGGCAGGACAGCCAGUUGGCUAGCAGUCAGUUGGCCAGCUAGGAGCCG______AG_____CAGGCCUUGCAGGACGA ........((..((((.........))))..)).((....(((((((((((((.....))))))))))).))..))............................... (-26.92 = -27.58 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:43 2006