
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,147,474 – 9,147,602 |
| Length | 128 |
| Max. P | 0.883864 |

| Location | 9,147,474 – 9,147,568 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 91.82 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -24.89 |
| Energy contribution | -25.25 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
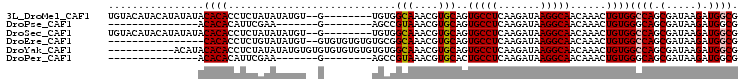
>3L_DroMel_CAF1 9147474 94 + 23771897 AACCGAGCCCAAAACGUGUCGCCUCAAGUGCCACGCCAUCUUAUCGCUGGCCACAGUUUGUUGCCUUAUCUUGAGGCACUGCACGUUUGCCACA---- ....(.((...((((((((.((((((((......((.........)).(((.((.....)).)))....))))))))...)))))))))))...---- ( -28.00) >DroPse_CAF1 2676 94 + 1 AGCCAAGCCAAAAACGUGUCGCCUCAAGUGCCACGCCAUCUUAUCGCUGCCCACAGUUUGUUGCCUUAUCUUGAGGCACUGCACGUUUACGGCU---- .....((((..((((((((.((((((((......((...(.....((((....))))..)..)).....))))))))...))))))))..))))---- ( -30.20) >DroSec_CAF1 2565 94 + 1 AACCGAGCCCAAAACGUGUCGCCCCAAGUGCCACGCCAUCUUAUCGCUGGCCACAGUUUGUUGCCUUAUCUUGAGGCACUGCACGUUUGCCACA---- ....(.((...((((((((.(......((((((.((.........))))).))).......((((((.....))))))).)))))))))))...---- ( -24.30) >DroEre_CAF1 14559 98 + 1 AGCCGAGCCCAAAACGUGUCGCCUCAAGUGCCACGCCAUCUUAUCGCUGGCCACAGUUUGUUGCCUUAUCUUGAGGCACUGCACGUUUGCCGCACACA ......((...((((((((.((((((((......((.........)).(((.((.....)).)))....))))))))...))))))))...))..... ( -30.50) >DroAna_CAF1 2689 94 + 1 AGCCAAGCCCAAAACGUGUCGCCUCAAGUGCCACGCCAUCUUAUCGCUGCCCACAGUUUGUUGCCUUAUCUUGAGGCGCUGCACGUUUGUAGUG---- ......((...(((((((((((((((((......((...(.....((((....))))..)..)).....)))))))))..))))))))...)).---- ( -27.90) >DroPer_CAF1 2665 94 + 1 AGCCAAGCCAAAAACGUGUCGCCUCAAGUGCCACGCCAUCUUAUCGCUGCCCACAGUUUGUUGCCUUAUCUUGAGGCAGUGCACGUUUACGGCU---- .....((((..((((((((.((((((((......((...(.....((((....))))..)..)).....))))))))...))))))))..))))---- ( -30.20) >consensus AGCCAAGCCCAAAACGUGUCGCCUCAAGUGCCACGCCAUCUUAUCGCUGCCCACAGUUUGUUGCCUUAUCUUGAGGCACUGCACGUUUGCCGCA____ ......((...((((((((.((((((((......((...(.....((((....))))..)..)).....))))))))...))))))))...))..... (-24.89 = -25.25 + 0.36)



| Location | 9,147,507 – 9,147,602 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.69 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -14.29 |
| Energy contribution | -14.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
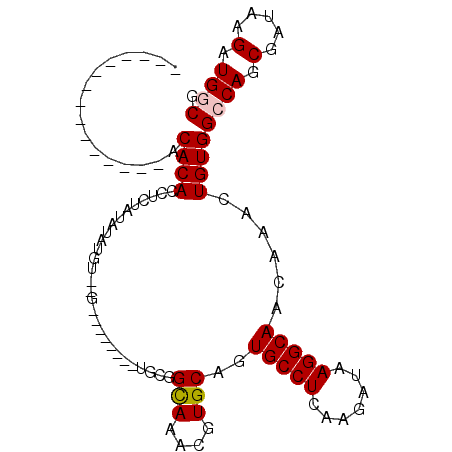
>3L_DroMel_CAF1 9147507 95 - 23771897 UGUACAUACAUAUAUACACACCUCUAUAUAUGU--G--------UGUGGCAAACGUGCAGUGCCUCAAGAUAAGGCAACAAACUGUGGCCAGCGAUAAGAUGGCG ((((((((((((((((........)))))))))--)--------))).))).....(((((((((.......)))).....))))).((((.(.....).)))). ( -28.80) >DroPse_CAF1 2709 75 - 1 ---------------ACACACAUUCGAA-------G--------AGCCGUAAACGUGCAGUGCCUCAAGAUAAGGCAACAAACUGUGGGCAGCGAUAAGAUGGCG ---------------.............-------.--------.(((((.....(((..((((.((......(....)......))))))))).....))))). ( -18.60) >DroSec_CAF1 2598 95 - 1 UGUACAUACAUAUAUACACACCUCUAUAUAUGU--G--------UGUGGCAAACGUGCAGUGCCUCAAGAUAAGGCAACAAACUGUGGCCAGCGAUAAGAUGGCG ((((((((((((((((........)))))))))--)--------))).))).....(((((((((.......)))).....))))).((((.(.....).)))). ( -28.80) >DroEre_CAF1 14592 87 - 1 ----------------CACACCUCUGUAUAUGU--GUGUGUGUGUGCGGCAAACGUGCAGUGCCUCAAGAUAAGGCAACAAACUGUGGCCAGCGAUAAGAUGGCG ----------------((((...(((((((..(--....)..))))))).....((....(((((.......)))))....))))))((((.(.....).)))). ( -23.70) >DroYak_CAF1 15735 94 - 1 -----------ACAUACACACCUCUAUAUAUGUGUGUGUGUGUGUGUGGCAAACGUGCAGUGCCUCAAGAUAAGGCAACAAACUGUGGCCAGCGAUAAGAUGGCG -----------(((((((((((.(.......).).)))))))))).((((....((....(((((.......)))))....))....))))((.((...)).)). ( -28.80) >DroPer_CAF1 2698 75 - 1 ---------------ACACACAUUCGAA-------G--------AGCCGUAAACGUGCACUGCCUCAAGAUAAGGCAACAAACUGUGGGCAGCGAUAAGAUGGCG ---------------.............-------.--------.(((((.......(.(((((.((......(....)......))))))).).....))))). ( -19.10) >consensus _______________ACACACCUCUAUAUAUGU__G________UGCGGCAAACGUGCAGUGCCUCAAGAUAAGGCAACAAACUGUGGCCAGCGAUAAGAUGGCG ................((((............................(((....)))..(((((.......)))))......))))((((.(.....).)))). (-14.29 = -14.40 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:38 2006