
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
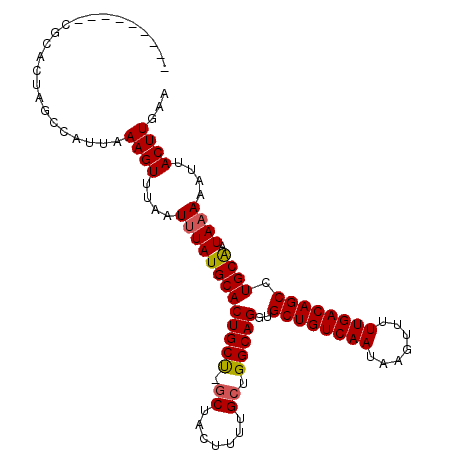
| Location | 9,146,529 – 9,146,627 |
| Length | 98 |
| Max. P | 0.999841 |

| Location | 9,146,529 – 9,146,627 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.07 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -26.83 |
| Energy contribution | -26.80 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.22 |
| SVM RNA-class probability | 0.999841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9146529 98 + 23771897 ---------CGCGCUAGCCAUUAAAGUUUAAUUUAUGCACUGCU-GCUACUUUUGCUGGCAGGUGCUGUCAAUAAGUUUUUGACAGCCUGCAAUAAAAAUUACUUGAA ---------.((((((((....(((((.........(((....)-)).))))).)))))).((.((((((((.......))))))))))))................. ( -27.10) >DroPse_CAF1 1964 98 + 1 ---------UCCACUAGCCAUUAAAGUUUAAUUUAUGCACUGCC-GCUACUUUUGCUGGCAGGUGCUGUCAAUAAGUUUUUGACAGCUUGCAAUAAAAAUUACUUGAA ---------..............((((....(((((((((((((-((.......)).)))))..((((((((.......)))))))).))).)))))....))))... ( -28.70) >DroSim_CAF1 1622 98 + 1 ---------CGCUCUAGCCAUUAAAGUUUAAUUUAUGCACUGCU-GCUACUUUUGCUGGCAGGUGCUGUCAAUAAGUUUUUGACAGCCUGCAAUAAAAAUUACUUGAA ---------.((....)).....((((....(((((((((((((-((.......)).)))))..((((((((.......)))))))).))).)))))....))))... ( -26.60) >DroEre_CAF1 13561 98 + 1 ---------CUUGCUAGCCAUUAAAGUUUAAUUUAUGCACUGCU-GCUACUUUUGCUGGCAGGUGCUGUCAAUAAGUUUUUGACAGCCUGCAAUAAAAAUUACUUGAA ---------(((((((((....(((((.........(((....)-)).))))).))))))))).((((((((.......))))))))..................... ( -26.70) >DroAna_CAF1 1748 107 + 1 UACGGUCUACACUCUAGCCAUUAAAGUUUAAUUUAUGCACUGCU-GCUACUUUGGCUGGCAGGUGCUGUCAAUAAGUUUUUGACAGCCUGCAAUAAAAAUUACUUGAA ...((.(((.....)))))....((((....(((((((((((((-(((.....))).)))))..((((((((.......)))))))).))).)))))....))))... ( -29.80) >DroPer_CAF1 1956 99 + 1 ---------CUCAGUAGGCAUGUAAGUUUAAUAUAUGCACUGCCCCCUACUUUUGCUGGCAGGUGCUGUCAAUAACUUGUUGACAGCUUGCGAUAAAAAUUACUUGAA ---------.((((((((((((((.......))))))).(((((..(.......)..)))))..(((((((((.....)))))))))............)))).))). ( -29.60) >consensus _________CGCACUAGCCAUUAAAGUUUAAUUUAUGCACUGCU_GCUACUUUUGCUGGCAGGUGCUGUCAAUAAGUUUUUGACAGCCUGCAAUAAAAAUUACUUGAA .......................((((....(((((((((((((.((.......)).)))))..((((((((.......)))))))).)))).))))....))))... (-26.83 = -26.80 + -0.03)


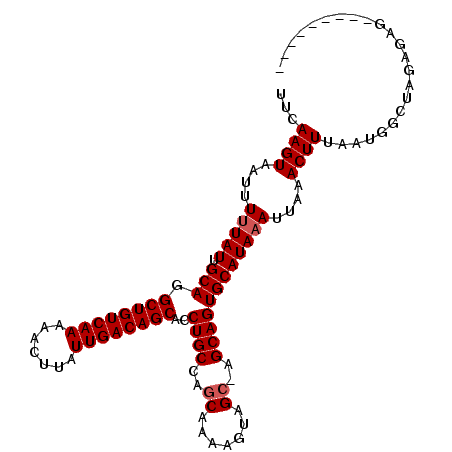
| Location | 9,146,529 – 9,146,627 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.07 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.58 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9146529 98 - 23771897 UUCAAGUAAUUUUUAUUGCAGGCUGUCAAAAACUUAUUGACAGCACCUGCCAGCAAAAGUAGC-AGCAGUGCAUAAAUUAAACUUUAAUGGCUAGCGCG--------- .....((.((((((.((((((((((((((.......))))))))..))).))).)))))).))-....((((....((((.....)))).....)))).--------- ( -23.70) >DroPse_CAF1 1964 98 - 1 UUCAAGUAAUUUUUAUUGCAAGCUGUCAAAAACUUAUUGACAGCACCUGCCAGCAAAAGUAGC-GGCAGUGCAUAAAUUAAACUUUAAUGGCUAGUGGA--------- ...((((....(((((.(((.((((((((.......))))))))..(((((.((.......))-)))))))))))))....))))..............--------- ( -26.90) >DroSim_CAF1 1622 98 - 1 UUCAAGUAAUUUUUAUUGCAGGCUGUCAAAAACUUAUUGACAGCACCUGCCAGCAAAAGUAGC-AGCAGUGCAUAAAUUAAACUUUAAUGGCUAGAGCG--------- ...((((....(((((.(((.((((((((.......))))))))..((((..((.......))-.))))))))))))....)))).....((....)).--------- ( -23.90) >DroEre_CAF1 13561 98 - 1 UUCAAGUAAUUUUUAUUGCAGGCUGUCAAAAACUUAUUGACAGCACCUGCCAGCAAAAGUAGC-AGCAGUGCAUAAAUUAAACUUUAAUGGCUAGCAAG--------- ...............((((.(((((((((.......))))).((((.(((..((.......))-.))))))).................)))).)))).--------- ( -23.30) >DroAna_CAF1 1748 107 - 1 UUCAAGUAAUUUUUAUUGCAGGCUGUCAAAAACUUAUUGACAGCACCUGCCAGCCAAAGUAGC-AGCAGUGCAUAAAUUAAACUUUAAUGGCUAGAGUGUAGACCGUA ...((((....(((((.(((.((((((((.......))))))))..((((..((.......))-.))))))))))))....))))..(((((((.....))).)))). ( -26.40) >DroPer_CAF1 1956 99 - 1 UUCAAGUAAUUUUUAUCGCAAGCUGUCAACAAGUUAUUGACAGCACCUGCCAGCAAAAGUAGGGGGCAGUGCAUAUAUUAAACUUACAUGCCUACUGAG--------- .(((.(((.........((..((((((((.......)))))))).(((((........)))))..)).(.((((.((.......)).))))))))))).--------- ( -24.80) >consensus UUCAAGUAAUUUUUAUUGCAGGCUGUCAAAAACUUAUUGACAGCACCUGCCAGCAAAAGUAGC_AGCAGUGCAUAAAUUAAACUUUAAUGGCUAGAGAG_________ ...((((....(((((.(((.((((((((.......))))))))..((((..((.......))..))))))))))))....))))....................... (-20.25 = -20.58 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:37 2006