| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,129,448 – 9,129,555 |
| Length | 107 |
| Max. P | 0.999729 |

| Location | 9,129,448 – 9,129,555 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.38 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.96 |
| SVM RNA-class probability | 0.999729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
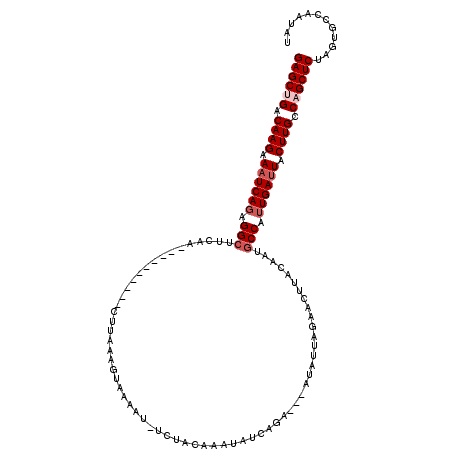
>3L_DroMel_CAF1 9129448 107 + 23771897 AUAUUGGCACUAGAGCUGGCAAGUAAUCAAUGGCAUUAUAAGUUAUAAUAU---UCUGAAAUUUGUUGAAAUUUUACUUUAAG----------UAGAUGCCUGUGAUUCCUUGUCAGCUC ............(((((((((((.(((((..((((((..(((((.(((((.---.........))))).)))))(((.....)----------))))))))..))))).))))))))))) ( -33.00) >DroSec_CAF1 4486 107 + 1 AUUUUGGCACUAGAGCUGGCAAGUAGUCAAUGGCAUUGUAUGUUCUAAUAU---UCUGAUUUUUGUAGAUAUUUUACUUUAAG----------UAGAAGCCUCUGAUUGCUUGUCAGCUC ............(((((((((((((((((..(((....(((......)))(---((((......(((((...)))))......----------))))))))..))))))))))))))))) ( -37.70) >DroSim_CAF1 4486 107 + 1 AUAUUGGCACUAGAGCUGGCAAGUAAUCAAUGGCAUUGUAAGUUCUAAUAU---UCAGAUUAUUGUAGAUAUUUAACUUUAAG----------UGGAAGCCUCUGAUUUCUUGUCAGCUC ............(((((((((((.(((((..((((((..(((((..(((((---..(.(....).)..))))).)))))..))----------)....)))..))))).))))))))))) ( -31.30) >DroEre_CAF1 4565 117 + 1 AUAUUGGCAAUAGAGCUGGCAAGUAAUCAAAGGCAUUCUAAGUUCGGAUAUAGUUUUUACAUUUUUGA--UUUUAAGU-UAAGUUUGAUUUGAUUUAAGCCCCUGAUUGCUUGUCGGCUC ............(((((((((((((((((..(((.........(((((.((.((....)))).)))))--..((((((-(((((...))))))))))))))..))))))))))))))))) ( -38.70) >DroYak_CAF1 4631 108 + 1 AUAUUGGCAAUAGAGCUGGCAAGUAAUCAAAGGUAUUCGAGGUUGUUAUAUAUAUUUAAGAUUUCUAG--GUUGCAGUUUAAG----------UUAAAGCCUCUGAUUUCUUGUCGGCUC ............(((((((((((.(((((.((((....((((((.(((........)))))))))(((--.(((.....))).----------)))..)))).))))).))))))))))) ( -29.60) >consensus AUAUUGGCACUAGAGCUGGCAAGUAAUCAAUGGCAUUCUAAGUUCUAAUAU___UCUGAUAUUUGUAGA_AUUUAACUUUAAG__________UAGAAGCCUCUGAUUGCUUGUCAGCUC ............(((((((((((.(((((..(((................................................................)))..))))).))))))))))) (-24.94 = -24.38 + -0.56)


| Location | 9,129,448 – 9,129,555 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.06 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -18.78 |
| Energy contribution | -19.78 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9129448 107 - 23771897 GAGCUGACAAGGAAUCACAGGCAUCUA----------CUUAAAGUAAAAUUUCAACAAAUUUCAGA---AUAUUAUAACUUAUAAUGCCAUUGAUUACUUGCCAGCUCUAGUGCCAAUAU ((((((.((((.(((((..(((((.((----------(.....)))((((((....))))))....---...............)))))..))))).)))).))))))............ ( -25.40) >DroSec_CAF1 4486 107 - 1 GAGCUGACAAGCAAUCAGAGGCUUCUA----------CUUAAAGUAAAAUAUCUACAAAAAUCAGA---AUAUUAGAACAUACAAUGCCAUUGACUACUUGCCAGCUCUAGUGCCAAAAU ((((((.((((...((((.(((...((----------(.....))).....((((...........---....)))).........))).))))...)))).))))))............ ( -22.26) >DroSim_CAF1 4486 107 - 1 GAGCUGACAAGAAAUCAGAGGCUUCCA----------CUUAAAGUUAAAUAUCUACAAUAAUCUGA---AUAUUAGAACUUACAAUGCCAUUGAUUACUUGCCAGCUCUAGUGCCAAUAU ((((((.((((.((((((.(((.....----------....(((((.((((((...........).---)))))..))))).....))).)))))).)))).))))))............ ( -26.14) >DroEre_CAF1 4565 117 - 1 GAGCCGACAAGCAAUCAGGGGCUUAAAUCAAAUCAAACUUA-ACUUAAAA--UCAAAAAUGUAAAAACUAUAUCCGAACUUAGAAUGCCUUUGAUUACUUGCCAGCUCUAUUGCCAAUAU ((((.(.((((.((((((((((...................-((......--........)).....(((..........)))...)))))))))).)))).).))))............ ( -22.74) >DroYak_CAF1 4631 108 - 1 GAGCCGACAAGAAAUCAGAGGCUUUAA----------CUUAAACUGCAAC--CUAGAAAUCUUAAAUAUAUAUAACAACCUCGAAUACCUUUGAUUACUUGCCAGCUCUAUUGCCAAUAU ((((.(.((((.(((((((((......----------......(((....--.)))...................(......)....))))))))).)))).).))))............ ( -19.40) >consensus GAGCUGACAAGAAAUCAGAGGCUUCAA__________CUUAAAGUAAAAU_UCUACAAAUAUCAGA___AUAUUAGAACUUACAAUGCCAUUGAUUACUUGCCAGCUCUAGUGCCAAUAU ((((((.((((.((((((.(((................................................................))).)))))).)))).))))))............ (-18.78 = -19.78 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:27 2006