| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,128,393 – 9,128,489 |
| Length | 96 |
| Max. P | 0.960096 |

| Location | 9,128,393 – 9,128,489 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -19.63 |
| Energy contribution | -18.47 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9128393 96 - 23771897 A------AAACUAUUUCGCAUUUCCUAUGGAGCCAAAGGACC--AGAGAA-AGUCUAUCCAUAGGAAUGCUGAAAUGCACUGCACCU-GAGGCACUGUUAAGGGGU .------..(((((((((((((.((((((((......((((.--......-.)))).))))))))))))).)))))(((.(((....-...))).))).....))) ( -26.90) >DroPse_CAF1 3747 103 - 1 ACCUACAUACAUAUUUCACAUU-AUUAUGGAGCCAACAGGCUUACGAGAA-AGUCUAUCUAUAAGGAUGCUGAAAUGCACUGCACUGAAAGGCACUGUAAUGG-GU (((((........(((((((((-.(((((((......((((((......)-))))).))))))).)))).)))))((((.(((........))).)))).)))-)) ( -25.50) >DroEre_CAF1 3521 95 - 1 A-------AACUGUUUCGCAUUUCUUAUGGAGCCAAAGGACC--AGAAAA-AGUCUAUCCAUAGGGAUGCCGAAAUGCACUGCACCU-GAGGCACUGUAAAGGGGU .-------.(((.(((..((.((((((((((......((((.--......-.)))).))))))))))((((....(((...)))...-..)))).))..))).))) ( -24.70) >DroYak_CAF1 3567 96 - 1 A------AAACUAUUUCGCAUUUCUUAUGGAGCCAAAGGACC--AGAAAA-AGUCCAUCCAUAGAGAUGCUGAAAUGCACUGCACCU-GAGGCACUGUAAAGGGGU .------.....((..(((((((((.(((((......((((.--......-.)))).))))))))))))).....((((.(((....-...))).))))..)..)) ( -27.30) >DroAna_CAF1 3909 87 - 1 --------AACUAUUUCACUUUUCUUAUGGAGCCAACAGACU--GCAAACUGGUCCAUCCAUAAGGACGCUGAAAUGCACUGCACUU-GAGGCACAGC-------- --------....((((((.(.((((((((((.....(((...--.....))).....)))))))))).).))))))((..(((.(..-..))))..))-------- ( -21.30) >DroPer_CAF1 3767 103 - 1 ACCUACAUACAUAUUUCACAUU-AUUAUGGAGCCAACAGGCUUACGAGAA-AGUCUAUCUAUAAGGAUGCUGAAAUGCACUGCACUGAAAGGCACUGUAAUGG-GU (((((........(((((((((-.(((((((......((((((......)-))))).))))))).)))).)))))((((.(((........))).)))).)))-)) ( -25.50) >consensus A_______AACUAUUUCACAUUUCUUAUGGAGCCAAAAGACC__AGAAAA_AGUCUAUCCAUAAGGAUGCUGAAAUGCACUGCACCU_GAGGCACUGUAAAGG_GU ............(((((((((((.(((((((......((((...........)))).)))))))))))).))))))(((.(((........))).)))........ (-19.63 = -18.47 + -1.16)


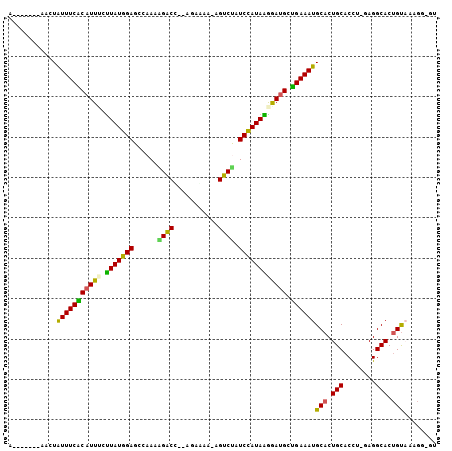
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:22 2006