
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,127,170 – 9,127,290 |
| Length | 120 |
| Max. P | 0.998067 |

| Location | 9,127,170 – 9,127,263 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.38 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -4.88 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9127170 93 + 23771897 UUA--UUUAUAUAUAUU-UGU-------UUGGCA-----AUUUGGCAGCCUUCAUGUCUGGGUCACCC-GUUUAAGUGACCCAAACUUGAGAGCUCCAAAUCGCAGAUA ...--............-(((-------(((.(.-----((((((.(((.((((.((.((((((((..-......)))))))).)).)))).))))))))).))))))) ( -33.40) >DroVir_CAF1 3609 90 + 1 -------GAGAGU--UU--------GCCCGGGCGUCAACAUUUGUAGCUUGUCAUGUCUGGGCCACGC-AUUUAAAUGGCGCAGACUUGACAGG-CCAAAUGUUAUAUA -------......--..--------((....))...((((((((..((((((((.(((((.((((...-.......)))).))))).)))))))-)))))))))..... ( -37.00) >DroSec_CAF1 2253 101 + 1 UUU-UUGUAUAUAUAUU-UUGUUUUUUUUUGGCA-----AUUUGGCAGCCUUCAUGUCUGGGUCACCC-CUUUAAGUGACCCAAACUUGAGAGCUCCAAAUCGCAGAUA ...-.............-.............((.-----((((((.(((.((((.((.((((((((..-......)))))))).)).)))).))))))))).))..... ( -32.80) >DroSim_CAF1 2254 100 + 1 UUU--UAUAUAUUUUUU-UGUUUUUUUUGUGGCG-----AUUUGGCAGCUUUCAUGUCUGGGUCACCC-CUUUAAGUGACCCAAACUUGAGAGCUCCAAAUCGCAGAUA ...--............-.............(((-----((((((.((((((((.((.((((((((..-......)))))))).)).)))))))))))))))))..... ( -43.20) >DroEre_CAF1 2281 103 + 1 UUUUUUGUACAUAUAUUGCUUUUUGUUUUUGGCA-----ACUUGGCAGCCCUCAUGUCUGGGUCACCC-CUUUAAAUGACCCAAACUUAAUGGCUCCAAAUCGCAGAUA ...(((((.......(((((..........))))-----).((((.((((.....((.(((((((...-.......))))))).)).....))))))))...))))).. ( -27.60) >DroWil_CAF1 2832 95 + 1 UCU--CGU----UUGUU-UGUAUUGUAUUUGAAA-----AUCUCAGAUCUAUUAUGUCUGGAUCACGUGCUUUAAGUGAUCCAAACUUAAUAGUUCGAAAUU-UAUAU- ..(--((.----.....-........((((((..-----...))))))((((((.((.((((((((.........)))))))).)).))))))..)))....-.....- ( -21.70) >consensus UUU__UGUAUAUAUAUU_UGUUUU_UUUUUGGCA_____AUUUGGCAGCCUUCAUGUCUGGGUCACCC_CUUUAAGUGACCCAAACUUGAGAGCUCCAAAUCGCAGAUA .......................................((((((.((((((((.((.((((((((.........)))))))).)).))))))))))))))........ (-23.98 = -24.18 + 0.20)

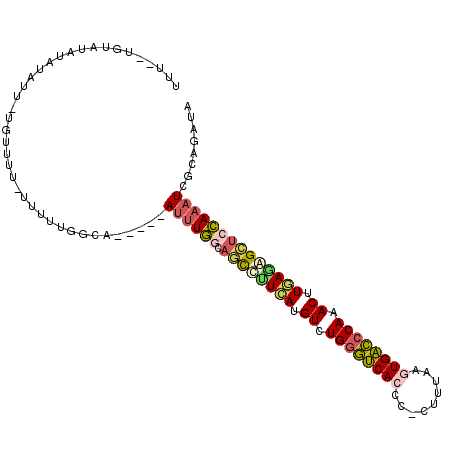

| Location | 9,127,170 – 9,127,263 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 67.38 |
| Mean single sequence MFE | -38.55 |
| Consensus MFE | -30.39 |
| Energy contribution | -28.87 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.54 |
| Mean z-score | -7.38 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9127170 93 - 23771897 UAUCUGCGAUUUGGAGCUCUCAAGUUUGGGUCACUUAAAC-GGGUGACCCAGACAUGAAGGCUGCCAAAU-----UGCCAA-------ACA-AAUAUAUAUAAA--UAA .....((((((((((((..(((.((((((((((((.....-.)))))))))))).)))..))).))))))-----)))...-------...-............--... ( -42.50) >DroVir_CAF1 3609 90 - 1 UAUAUAACAUUUGG-CCUGUCAAGUCUGCGCCAUUUAAAU-GCGUGGCCCAGACAUGACAAGCUACAAAUGUUGACGCCCGGGC--------AA--ACUCUC------- ....((((((((((-(.(((((.(((((.(((((......-..))))).))))).))))).))..)))))))))..((....))--------..--......------- ( -34.70) >DroSec_CAF1 2253 101 - 1 UAUCUGCGAUUUGGAGCUCUCAAGUUUGGGUCACUUAAAG-GGGUGACCCAGACAUGAAGGCUGCCAAAU-----UGCCAAAAAAAAACAA-AAUAUAUAUACAA-AAA .....((((((((((((..(((.(((((((((((((....-))))))))))))).)))..))).))))))-----))).............-.............-... ( -42.80) >DroSim_CAF1 2254 100 - 1 UAUCUGCGAUUUGGAGCUCUCAAGUUUGGGUCACUUAAAG-GGGUGACCCAGACAUGAAAGCUGCCAAAU-----CGCCACAAAAAAAACA-AAAAAAUAUAUA--AAA .....(((((((((((((.(((.(((((((((((((....-))))))))))))).))).)))).))))))-----))).............-............--... ( -44.50) >DroEre_CAF1 2281 103 - 1 UAUCUGCGAUUUGGAGCCAUUAAGUUUGGGUCAUUUAAAG-GGGUGACCCAGACAUGAGGGCUGCCAAGU-----UGCCAAAAACAAAAAGCAAUAUAUGUACAAAAAA .....(((((((((((((.(((.(((((((((((((....-))))))))))))).))).)))).))))))-----)))............................... ( -39.60) >DroWil_CAF1 2832 95 - 1 -AUAUA-AAUUUCGAACUAUUAAGUUUGGAUCACUUAAAGCACGUGAUCCAGACAUAAUAGAUCUGAGAU-----UUUCAAAUACAAUACA-AACAA----ACG--AGA -....(-((((((((.((((((.(((((((((((.........))))))))))).)))))).)).)))))-----))..............-.....----...--... ( -27.20) >consensus UAUCUGCGAUUUGGAGCUCUCAAGUUUGGGUCACUUAAAG_GGGUGACCCAGACAUGAAAGCUGCCAAAU_____UGCCAAAAA_AAAACA_AAUAAAUAUACA__AAA ........((((((((((.(((.(((((((((((((.....))))))))))))).))).)))).))))))....................................... (-30.39 = -28.87 + -1.52)
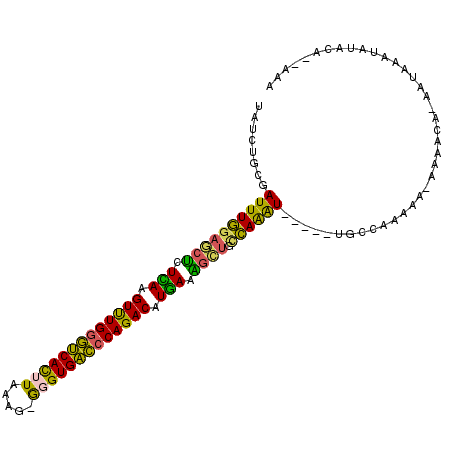
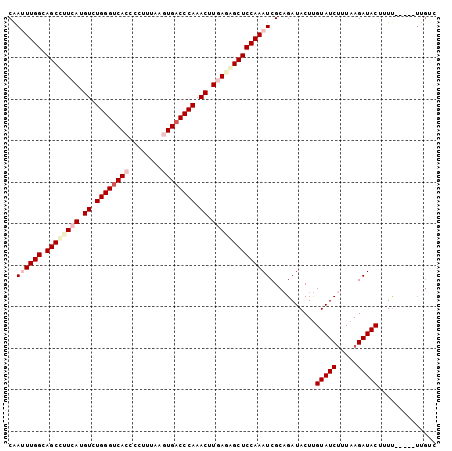
| Location | 9,127,192 – 9,127,290 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -31.48 |
| Energy contribution | -33.07 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.07 |
| Mean z-score | -5.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9127192 98 + 23771897 CAAUUUGGCAGCCUUCAUGUCUGGGUCACCCGUUUAAGUGACCCAAACUUGAGAGCUCCAAAUCGCAGAUACUUGUAUCUUUAAGAUACUAUU-----UUGCC ..((((((.(((.((((.((.((((((((........)))))))).)).)))).))))))))).(((((.....(((((.....)))))...)-----)))). ( -37.70) >DroSec_CAF1 2283 97 + 1 CAAUUUGGCAGCCUUCAUGUCUGGGUCACCCCUUUAAGUGACCCAAACUUGAGAGCUCCAAAUCGCAGAUACUUGUAUCUUUAAGAUACU-UU-----UGGUC ..((((((.(((.((((.((.((((((((........)))))))).)).)))).)))))))))..((((.....(((((.....))))).-))-----))... ( -34.00) >DroSim_CAF1 2283 94 + 1 CGAUUUGGCAGCUUUCAUGUCUGGGUCACCCCUUUAAGUGACCCAAACUUGAGAGCUCCAAAUCGCAGAUACUUGUAUCUUUAAGAUACU-UU-----UU--- ((((((((.((((((((.((.((((((((........)))))))).)).)))))))))))))))).........(((((.....))))).-..-----..--- ( -42.80) >DroEre_CAF1 2313 98 + 1 CAACUUGGCAGCCCUCAUGUCUGGGUCACCCCUUUAAAUGACCCAAACUUAAUGGCUCCAAAUCGCAGAUACUUGUAUCCUUAAGAUACUUUU-----AUGUC ....((((.((((.....((.(((((((..........))))))).)).....))))))))...(((.......(((((.....)))))....-----.))). ( -26.10) >DroYak_CAF1 2327 98 + 1 CAACUUGGCAGCCCUCAUGUCUGGGUCACCCCUUUAAAUGACCCAAACUUAAGGGCUCCAAAUCGCAGAUACUUGUAUCUGUAAGAUACUUUC-----UUGUC ....((((.((((((...((.(((((((..........))))))).))...))))))))))...(((((((....)))))))((((.....))-----))... ( -36.20) >DroAna_CAF1 2592 103 + 1 CAAUUUGGCAGCCAUCAUGUCUGGGACACGCGUUAAAGUGACCCAAACUUGAGGGCUCCAAAUCGCCGAUACUCGUAUCUCUCAGAUACUUUUCGACUUUGAC ..((((((.((((.(((.((.((((.(((........))).)))).)).))).))))))))))...(((.....(((((.....)))))...)))........ ( -33.20) >consensus CAAUUUGGCAGCCUUCAUGUCUGGGUCACCCCUUUAAGUGACCCAAACUUGAGAGCUCCAAAUCGCAGAUACUUGUAUCUUUAAGAUACUUUU_____UUGUC ..((((((.((((((((.((.((((((((........)))))))).)).))))))))))))))...........(((((.....))))).............. (-31.48 = -33.07 + 1.58)


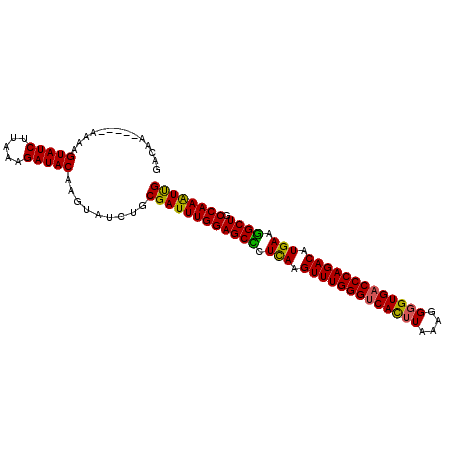
| Location | 9,127,192 – 9,127,290 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -42.66 |
| Energy contribution | -41.80 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.18 |
| Mean z-score | -7.00 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9127192 98 - 23771897 GGCAA-----AAUAGUAUCUUAAAGAUACAAGUAUCUGCGAUUUGGAGCUCUCAAGUUUGGGUCACUUAAACGGGUGACCCAGACAUGAAGGCUGCCAAAUUG .....-----....(((((.....))))).........(((((((((((..(((.((((((((((((......)))))))))))).)))..))).)))))))) ( -43.50) >DroSec_CAF1 2283 97 - 1 GACCA-----AA-AGUAUCUUAAAGAUACAAGUAUCUGCGAUUUGGAGCUCUCAAGUUUGGGUCACUUAAAGGGGUGACCCAGACAUGAAGGCUGCCAAAUUG .....-----..-.(((((.....))))).........(((((((((((..(((.(((((((((((((....))))))))))))).)))..))).)))))))) ( -43.80) >DroSim_CAF1 2283 94 - 1 ---AA-----AA-AGUAUCUUAAAGAUACAAGUAUCUGCGAUUUGGAGCUCUCAAGUUUGGGUCACUUAAAGGGGUGACCCAGACAUGAAAGCUGCCAAAUCG ---..-----..-.(((((.....))))).........((((((((((((.(((.(((((((((((((....))))))))))))).))).)))).)))))))) ( -45.50) >DroEre_CAF1 2313 98 - 1 GACAU-----AAAAGUAUCUUAAGGAUACAAGUAUCUGCGAUUUGGAGCCAUUAAGUUUGGGUCAUUUAAAGGGGUGACCCAGACAUGAGGGCUGCCAAGUUG .....-----....(((((.....))))).........((((((((((((.(((.(((((((((((((....))))))))))))).))).)))).)))))))) ( -40.70) >DroYak_CAF1 2327 98 - 1 GACAA-----GAAAGUAUCUUACAGAUACAAGUAUCUGCGAUUUGGAGCCCUUAAGUUUGGGUCAUUUAAAGGGGUGACCCAGACAUGAGGGCUGCCAAGUUG ...((-----((.....)))).((((((....))))))((((((((((((((((.(((((((((((((....))))))))))))).)))))))).)))))))) ( -48.50) >DroAna_CAF1 2592 103 - 1 GUCAAAGUCGAAAAGUAUCUGAGAGAUACGAGUAUCGGCGAUUUGGAGCCCUCAAGUUUGGGUCACUUUAACGCGUGUCCCAGACAUGAUGGCUGCCAAAUUG ......(((((.(.(((((.....)))))...).)))))(((((((((((.(((.(((((((.(((........))).))))))).))).)))).))))))). ( -44.30) >consensus GACAA_____AAAAGUAUCUUAAAGAUACAAGUAUCUGCGAUUUGGAGCCCUCAAGUUUGGGUCACUUAAAGGGGUGACCCAGACAUGAAGGCUGCCAAAUUG ..............(((((.....))))).........((((((((((((.(((.(((((((((((((....))))))))))))).))).)))).)))))))) (-42.66 = -41.80 + -0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:20 2006