
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,093,429 – 9,093,559 |
| Length | 130 |
| Max. P | 0.882129 |

| Location | 9,093,429 – 9,093,532 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -26.63 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9093429 103 + 23771897 CUUUAACUGAUUGUGUGUGUGUGUAGAUUGUAUGUGUAUGUGUGUAUAAAUAGUUGG-GGCAUGAAGACGCUGCUAACUAUUUCUGUAGACGCAGCUGAAUGUG (((((((((.(((((..((..((((.((...)).))))..))..))))).)))))))-)).........(((((...((((....))))..)))))........ ( -23.40) >DroSec_CAF1 12730 87 + 1 CUUUCACUGAUUGUGUGUGUG-----------------UGUGUGUAUACAUAGUUGGGGGCGUGAAGAAGCUGCCAACUAUUUCUGUAGACGCAGCUGAAUGUG ....(((.....)))......-----------------(((((.((((.((((((((.(((........))).))))))))...)))).))))).......... ( -26.20) >DroSim_CAF1 16808 85 + 1 CUUUAACUUAUUGUGUGUG-------------------UGUCUGUAUACAUAGUUGGGGGCGUGAAGACGCUGCCAACUAUUUCUGUAGACGCAGCUGAAUGUG ........((((..((.((-------------------((((((((.(.((((((((.(((((....))))).)))))))).).))))))))))))..)))).. ( -30.30) >consensus CUUUAACUGAUUGUGUGUGUG_________________UGUGUGUAUACAUAGUUGGGGGCGUGAAGACGCUGCCAACUAUUUCUGUAGACGCAGCUGAAUGUG ......................................(((((.((((.((((((((.(((........))).))))))))...)))).))))).......... (-21.62 = -21.73 + 0.11)

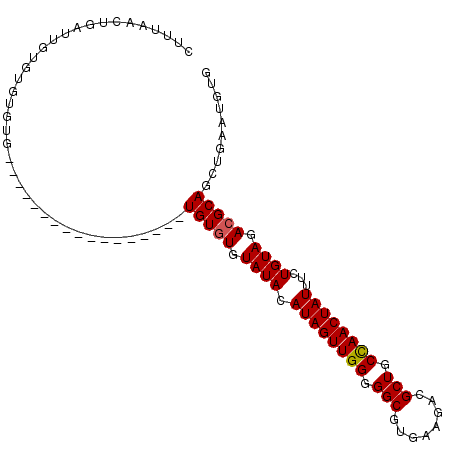
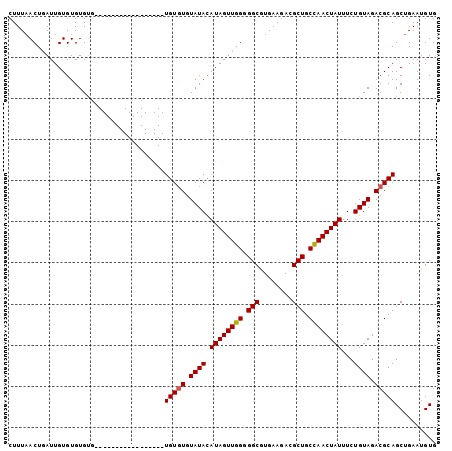
| Location | 9,093,429 – 9,093,532 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -15.90 |
| Consensus MFE | -10.71 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9093429 103 - 23771897 CACAUUCAGCUGCGUCUACAGAAAUAGUUAGCAGCGUCUUCAUGCC-CCAACUAUUUAUACACACAUACACAUACAAUCUACACACACACACAAUCAGUUAAAG ........(((((..(((......)))...)))))...........-......................................................... ( -10.50) >DroSec_CAF1 12730 87 - 1 CACAUUCAGCUGCGUCUACAGAAAUAGUUGGCAGCUUCUUCACGCCCCCAACUAUGUAUACACACA-----------------CACACACACAAUCAGUGAAAG ........(.((.((.((((....(((((((..((........))..))))))))))).)).))).-----------------......(((.....))).... ( -16.20) >DroSim_CAF1 16808 85 - 1 CACAUUCAGCUGCGUCUACAGAAAUAGUUGGCAGCGUCUUCACGCCCCCAACUAUGUAUACAGACA-------------------CACACACAAUAAGUUAAAG ........(.((.((((......((((((((..((((....))))..))))))))......)))).-------------------)))................ ( -21.00) >consensus CACAUUCAGCUGCGUCUACAGAAAUAGUUGGCAGCGUCUUCACGCCCCCAACUAUGUAUACACACA_________________CACACACACAAUCAGUUAAAG .........(((......)))..((((((((..((((....))))..))))))))................................................. (-10.71 = -11.27 + 0.56)



| Location | 9,093,462 – 9,093,559 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.41 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -14.94 |
| Energy contribution | -16.62 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9093462 97 - 23771897 UGGCAACAAAGGAGGCAGCAGCAGUGGCACAUUCAGCUGCGUCUACAGAAAUAGUUAGCAGCGUCUUCAUGCC-CCAACUAUUUAUACACACAUACAC .((((.....((((((....((....)).......(((((..(((......)))...))))))))))).))))-........................ ( -22.80) >DroSec_CAF1 12751 93 - 1 UGGCAACAAAGGAGGCAGCAGCAGUGGCACAUUCAGCUGCGUCUACAGAAAUAGUUGGCAGCUUCUUCACGCCCCCAACUAUGUAUACACACA----- ((....))..(((.(((((.(.((((...))))).))))).)))......((((((((..((........))..))))))))...........----- ( -26.30) >DroSim_CAF1 16827 93 - 1 UGGCAACAAAGGAGGCAGCAGCAGUGGCACAUUCAGCUGCGUCUACAGAAAUAGUUGGCAGCGUCUUCACGCCCCCAACUAUGUAUACAGACA----- ((....))......(((((.(.((((...))))).)))))((((......((((((((..((((....))))..))))))))......)))).----- ( -30.50) >DroEre_CAF1 12486 96 - 1 UGGCAACAAAGGAGGCAGCAGCAGUAGCACAUUCAGCUGCGUCUACAGAAAUAGUUGGCAGCGUCUUCACGCCCCCAACUAUGUAUACACAUA--CAC ((....))..(((.(((((................))))).)))......((((((((..((((....))))..))))))))((((....)))--).. ( -29.19) >DroWil_CAF1 14751 85 - 1 -----ACAAAGGCAACAGCAGCAGCUGCU---GGUGCUACGUUUGUAAAAAUAGUUGCUCCAUCCUUCAUCCGCCCUAACAUUUCACCAC-----CAC -----.....(((..((((....)))).(---((.((.((((((....)))).)).)).)))..........)))...............-----... ( -16.50) >consensus UGGCAACAAAGGAGGCAGCAGCAGUGGCACAUUCAGCUGCGUCUACAGAAAUAGUUGGCAGCGUCUUCACGCCCCCAACUAUGUAUACACACA__CAC ..........(.((((.(((((.............))))))))).)....((((((((..((((....))))..))))))))................ (-14.94 = -16.62 + 1.68)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:09 2006