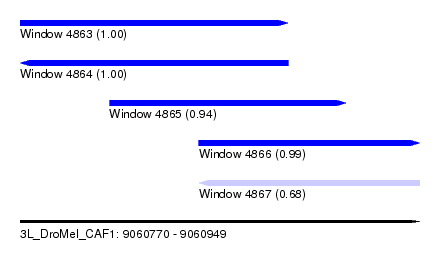
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,060,770 – 9,060,949 |
| Length | 179 |
| Max. P | 0.999879 |

| Location | 9,060,770 – 9,060,890 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.22 |
| Mean single sequence MFE | -38.23 |
| Consensus MFE | -24.42 |
| Energy contribution | -26.05 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.64 |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9060770 120 + 23771897 UAAACGGAGGCUAUCAGCCUCCCAAUGAGUUAAAUGGAGCCUACCCGGCACCUUCUCAGUUAAACGGAGGCUACCAGCCUCCUUUAGAAGGGAAUUUAGGUUACCGACCACUUUCUCAAC .....((((((.....))))))...((((..((.(((.(((.....)))((((((((..(((((.((((((.....)))))))))))..))))....))))......))).)).)))).. ( -43.60) >DroSec_CAF1 6200 120 + 1 UAAACGGAGCCUACCAACCUCCUUCUCAGUUGAACGGAGGCUACAAGCCACCUUCUCAGUUUAACGAAGGCUACCAGCCUCCUGUAGAAAGAGAUUGGGGUUACCAGCCACCUUCUCAAC .....((((.........))))......(((((..((.((((...((((.((.((((..((((....((((.....))))....))))..))))..))))))...)))).))...))))) ( -35.50) >DroEre_CAF1 6200 120 + 1 UAAAUGGAGGCUACCAGCCACCUUCUCAGCCAAACGGAGGCUACCAGCCACCCUCUCAGCCAAACGGAGGCUAUCAGCCACCCGUGGACAGAGAUUGGGGUUACCAGCCACCAUCUCAAG ...((((.((((...((((.((.((((..(((...((.((((...((((.((.............)).))))...)))).))..)))...))))..))))))...)))).))))...... ( -41.92) >DroYak_CAF1 6109 120 + 1 CAAACGGAGGUUAUCAGCCACCUUCUCAGUCAAGCGGAGGUUACCAGCCACCUGUAGAAAGAGAUUGGGGUUACCAGCCACCAUCUCAAGAAAAUAGUGGAUACCAGCCACCACACCAAU .....((.((((.....((((.(((((((....((((......)).))...))).)))).(((((.(((((.....))).))))))).........)))).....)))).))........ ( -31.90) >consensus UAAACGGAGGCUACCAGCCACCUUCUCAGUCAAACGGAGGCUACCAGCCACCUUCUCAGUUAAACGGAGGCUACCAGCCACCUGUAGAAAGAAAUUGGGGUUACCAGCCACCAUCUCAAC .....((.((((...((((((..((((........((.(((.....))).))........((((.((((((.....)))))).))))...))))..))))))...)))).))........ (-24.42 = -26.05 + 1.63)



| Location | 9,060,770 – 9,060,890 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.22 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -27.31 |
| Energy contribution | -28.87 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9060770 120 - 23771897 GUUGAGAAAGUGGUCGGUAACCUAAAUUCCCUUCUAAAGGAGGCUGGUAGCCUCCGUUUAACUGAGAAGGUGCCGGGUAGGCUCCAUUUAACUCAUUGGGAGGCUGAUAGCCUCCGUUUA ..((((.((((((..(((.((((......((((((...((((((.....)))))).........))))))....))))..)))))))))..))))...((((((.....))))))..... ( -43.10) >DroSec_CAF1 6200 120 - 1 GUUGAGAAGGUGGCUGGUAACCCCAAUCUCUUUCUACAGGAGGCUGGUAGCCUUCGUUAAACUGAGAAGGUGGCUUGUAGCCUCCGUUCAACUGAGAAGGAGGUUGGUAGGCUCCGUUUA .....(((.(..(((.......(((((((((((((.((((((((((((.((((((..........)))))).)))...)))))))(.....))))))))))))))))..)))..).))). ( -46.50) >DroEre_CAF1 6200 120 - 1 CUUGAGAUGGUGGCUGGUAACCCCAAUCUCUGUCCACGGGUGGCUGAUAGCCUCCGUUUGGCUGAGAGGGUGGCUGGUAGCCUCCGUUUGGCUGAGAAGGUGGCUGGUAGCCUCCAUUUA ....((((((.(((((.((.((((..((((.(((.(((((.(((((.(((((.((.(((.....))).)).))))).))))))))))..))).)))).)).)).)).))))).)))))). ( -57.00) >DroYak_CAF1 6109 120 - 1 AUUGGUGUGGUGGCUGGUAUCCACUAUUUUCUUGAGAUGGUGGCUGGUAACCCCAAUCUCUUUCUACAGGUGGCUGGUAACCUCCGCUUGACUGAGAAGGUGGCUGAUAACCUCCGUUUG ...(((..(((.((..(...((((((((((...)))))))))))..)).))).((..(.((((((.(((((((..((...)).)))))))....)))))).)..))...)))........ ( -37.40) >consensus GUUGAGAAGGUGGCUGGUAACCCCAAUCUCCUUCUAAAGGAGGCUGGUAGCCUCCGUUUAACUGAGAAGGUGGCUGGUAGCCUCCGUUUAACUGAGAAGGAGGCUGAUAGCCUCCGUUUA .....(((((.((.(((.....))).)).)))))....((((((((.((((((((.............((.(((.....))).)).............)))))))).))))))))..... (-27.31 = -28.87 + 1.56)

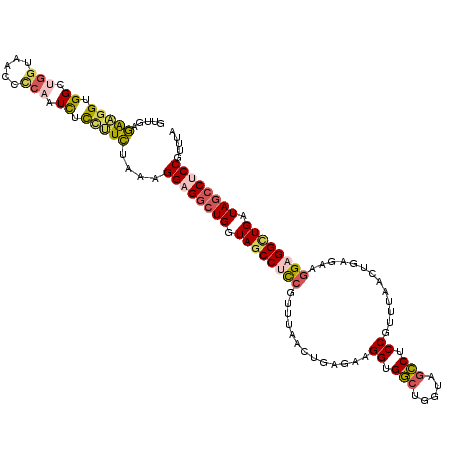
| Location | 9,060,810 – 9,060,916 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.11 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -15.24 |
| Energy contribution | -16.62 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9060810 106 + 23771897 CUACCCGGCACCUUCUCAGUUAAACGGAGGCUACCAGCCUCCUUUAGAAGGGAAUUUAGGUUACCGACCACUUUCUCAACAAAAUAGUGGAUAUCAGCCACCACAG ......(((((((((((..(((((.((((((.....)))))))))))..))))....))))......(((((.............)))))......)))....... ( -32.02) >DroSec_CAF1 6240 106 + 1 CUACAAGCCACCUUCUCAGUUUAACGAAGGCUACCAGCCUCCUGUAGAAAGAGAUUGGGGUUACCAGCCACCUUCUCAACAAAAUAGUGGAUAUCAGCCAGCACAG ......((..((.((((..((((....((((.....))))....))))..))))..))((((.....((((...............)))).....)))).)).... ( -24.06) >DroEre_CAF1 6240 106 + 1 CUACCAGCCACCCUCUCAGCCAAACGGAGGCUAUCAGCCACCCGUGGACAGAGAUUGGGGUUACCAGCCACCAUCUCAAGAAAAUAGUGGAUAUCAGCCACCACAC .......((((.......((((...((.(((.....))).))..))).).(((((.(((((.....))).))))))).........))))................ ( -27.80) >DroYak_CAF1 6149 106 + 1 UUACCAGCCACCUGUAGAAAGAGAUUGGGGUUACCAGCCACCAUCUCAAGAAAAUAGUGGAUACCAGCCACCACACCAAUCAACUGGUGGAUAUCAGCCACCUAUC .............((((...(((((.(((((.....))).))))))).........((((.......((((((...........)))))).......)))))))). ( -27.74) >consensus CUACCAGCCACCUUCUCAGUUAAACGGAGGCUACCAGCCACCUGUAGAAAGAAAUUGGGGUUACCAGCCACCAUCUCAACAAAAUAGUGGAUAUCAGCCACCACAC ............((((...(((((.((((((.....)))))).))))).)))).....((((.....(((((.............))))).....))))....... (-15.24 = -16.62 + 1.38)

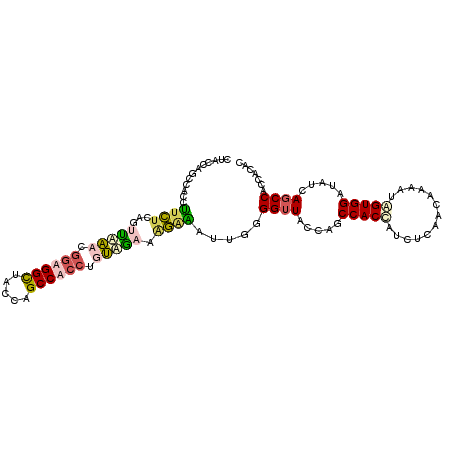
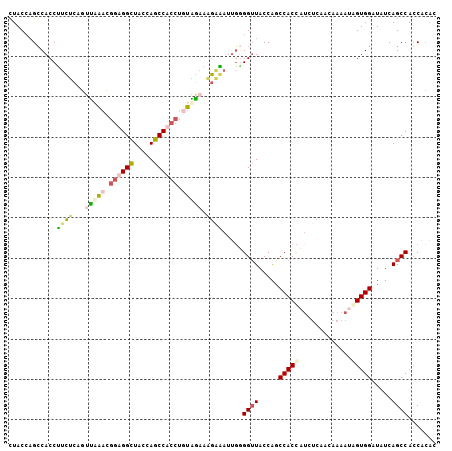
| Location | 9,060,850 – 9,060,949 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -14.87 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9060850 99 + 23771897 CCUUUAGAAGGGAAUUUAGGUUACCGACCACUUUCUCAACAAAAUAGUGGAUAUCAGCCACCACAGCCAUCAACUGGUGGAUAUCAGCCACCUUUCCAA ......((((((......((((.....(((((.............))))).....)))).......(((((....)))))..........))))))... ( -21.92) >DroSec_CAF1 6280 99 + 1 CCUGUAGAAAGAGAUUGGGGUUACCAGCCACCUUCUCAACAAAAUAGUGGAUAUCAGCCAGCACAGUCAUUAACUGGUGGAUAUCAGCCACCUUCCCAA ......(((.((((..(((((.....))).)).)))).........(((((((((.(((((............))))).)))))...)))).))).... ( -24.60) >DroEre_CAF1 6280 99 + 1 CCCGUGGACAGAGAUUGGGGUUACCAGCCACCAUCUCAAGAAAAUAGUGGAUAUCAGCCACCACACCAACCAACUGGUGGCUACCAGCCACCCCUCCAA ....((((..(((((.(((((.....))).))))))).........((((.....((((((((...........)))))))).....))))...)))). ( -34.20) >DroYak_CAF1 6189 99 + 1 CCAUCUCAAGAAAAUAGUGGAUACCAGCCACCACACCAAUCAACUGGUGGAUAUCAGCCACCUAUCCAAUCAACUGGUGGAUAUCAGCCACCUAUCCAA .................((((((...((.....(((((......)))))((((((.((((..............)))).)))))).))....)))))). ( -23.84) >consensus CCUGUAGAAAGAAAUUGGGGUUACCAGCCACCAUCUCAACAAAAUAGUGGAUAUCAGCCACCACACCAAUCAACUGGUGGAUAUCAGCCACCUUUCCAA ..................((((.....(((((.............))))).....))))................(((((.......)))))....... (-14.87 = -15.50 + 0.63)



| Location | 9,060,850 – 9,060,949 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9060850 99 - 23771897 UUGGAAAGGUGGCUGAUAUCCACCAGUUGAUGGCUGUGGUGGCUGAUAUCCACUAUUUUGUUGAGAAAGUGGUCGGUAACCUAAAUUCCCUUCUAAAGG ..((((((((.(((((...((((...(..(..(..(((((((.......))))))).)..)..)....))))))))).))))...)))).......... ( -27.30) >DroSec_CAF1 6280 99 - 1 UUGGGAAGGUGGCUGAUAUCCACCAGUUAAUGACUGUGCUGGCUGAUAUCCACUAUUUUGUUGAGAAGGUGGCUGGUAACCCCAAUCUCUUUCUACAGG (..(.(((((((..((((((..(((((..........)))))..))))))..))))))).)..)(((((.(..(((.....)))..).)))))...... ( -29.60) >DroEre_CAF1 6280 99 - 1 UUGGAGGGGUGGCUGGUAGCCACCAGUUGGUUGGUGUGGUGGCUGAUAUCCACUAUUUUCUUGAGAUGGUGGCUGGUAACCCCAAUCUCUGUCCACGGG .....(((((.((..((((((((((...........)))))))))....((((((((((...)))))))))))..)).))))).....(((....))). ( -39.30) >DroYak_CAF1 6189 99 - 1 UUGGAUAGGUGGCUGAUAUCCACCAGUUGAUUGGAUAGGUGGCUGAUAUCCACCAGUUGAUUGGUGUGGUGGCUGGUAUCCACUAUUUUCUUGAGAUGG .((((((...((((...((((((((((..(((((...((((.....))))..)))))..))))))).)))))))..))))))(((((((...))))))) ( -33.10) >consensus UUGGAAAGGUGGCUGAUAUCCACCAGUUGAUGGCUGUGGUGGCUGAUAUCCACUAUUUUGUUGAGAAGGUGGCUGGUAACCCCAAUCUCCUUCUAAAGG ..((((((((((.......))))).............(((.((((....(((((((((.....))))))))).)))).)))....)))))......... (-19.98 = -20.60 + 0.62)

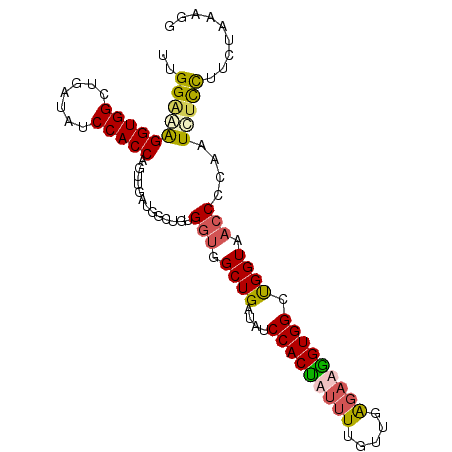

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:04 2006