
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,060,242 – 9,060,450 |
| Length | 208 |
| Max. P | 0.976157 |

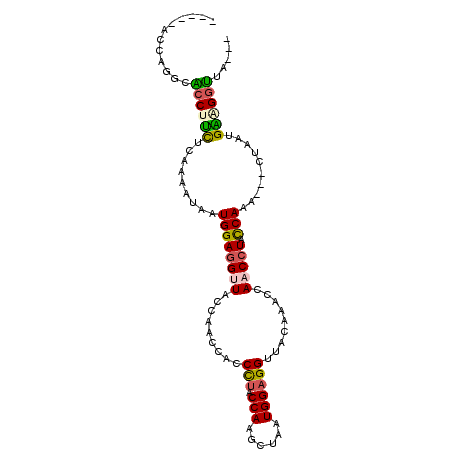
| Location | 9,060,242 – 9,060,336 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 69.24 |
| Mean single sequence MFE | -21.88 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.96 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9060242 94 + 23771897 -----AUCAGGCACCUUUUCAAAAUAAUGGAGGUUACCAACCAUCCUACCAAGGCAAUGGAGGUUACACACCAACAUACCAAAACCCUUUUGAGGGUUA--- -----....((.((((((..........))))))..))..(((((((....)))..)))).(((.....)))..........((((((....)))))).--- ( -19.80) >DroSec_CAF1 5667 99 + 1 AGGUUACCAGGCACCUUCUCAAAAUAAUGGAGGUUACCAACCAACCUACCAAGGUAAUGGAGGUUACACACCAACCUAUCAAAACACUUUUGAUGGUUA--- .(((.(((.((.((((((..........))))))..))..(((((((....))))..))).))).....)))((((.((((((.....)))))))))).--- ( -26.30) >DroEre_CAF1 5685 96 + 1 AGGCUACCA---GCCACCCCAAGCUAAUGGAGGCUACCAGCCACCCCACCAACCUAAUGGAGGCUACCAACCACCCUACCAAG---CUAAUGGAGGCUACAG .(((.....---))).(((((((((..(((.(((.....)))...((.(((......))).))...............)))))---))..))).))...... ( -24.80) >DroYak_CAF1 5605 86 + 1 --------AGCCGACUUCUCAAAACAAUGGAGGUUACCAGACACCUUACCAAACUAAUGGAGGUUACCAAGAACCCUACCAAG---CUAAUGGAGGU----- --------((((((((((.((......)))))))).............(((......))).)))).........(((.(((..---....)))))).----- ( -16.60) >consensus _____ACCAGGCACCUUCUCAAAAUAAUGGAGGUUACCAACCACCCUACCAAGCUAAUGGAGGUUACAAACCAACCUACCAAA___CUAAUGAAGGUUA___ ............((((((.........((((((((.........(((.(((......)))))).........))))).)))..........))))))..... (-11.70 = -11.96 + 0.25)

| Location | 9,060,336 – 9,060,450 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -12.35 |
| Energy contribution | -12.97 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9060336 114 + 23771897 CCACCAACCCAAUCAAUCAUCGUAUCAAACAAAUGGUGGAUACCAGCCACCCUUACCACCUCAUGGAGGUUACCAACCACCUUCUCAAACUUAUGGAGGUUUCCAACAACCUAA ...(((............................(((((.......))))).............((((((........)))))).........)))(((((......))))).. ( -20.00) >DroSec_CAF1 5766 114 + 1 CCACCAACCCAAUCAAUCUUCGCUUCAAACAAAUGGAGGAUAUCAGCCACCCUUACCACCUCAUGGAGGUUACCAACCACCUUCUCAAACUUAUGGAGGUUUCCAACAACCUAA ...(((.............((.(((((......)))))))........................((((((........)))))).........)))(((((......))))).. ( -17.00) >DroEre_CAF1 5781 114 + 1 CCACCAACCCAAUCAAUCAGCAGUUCAAUCAAAUGGAGGAUACCAGCCACCUUUACCAGCCUAUGGAGGUUACCAACCACCUUCUCAACCGUUUGGAGGCUUCCAGCAGCCUAA ............................((((((((.((.......))................((((((........))))))....))))))))(((((......))))).. ( -22.00) >DroYak_CAF1 5691 101 + 1 -CACCAGCCAUAU------------CAAGCAAACGGAGGAUACCAGCCACCCUUACCAGCCUAUGGAGGUUACCAACAACCUGCUCAAUCAUAUGGAGCUGUCCACCAACCCAA -.....((.....------------...))....((.(((((.............(((.....)))(((((......)))))((((.........))))))))).))....... ( -19.40) >consensus CCACCAACCCAAUCAAUCAUCG_UUCAAACAAAUGGAGGAUACCAGCCACCCUUACCACCCCAUGGAGGUUACCAACCACCUUCUCAAACUUAUGGAGGUUUCCAACAACCUAA ...(((............................((.((.......)).)).............((((((........)))))).........)))(((((......))))).. (-12.35 = -12.97 + 0.62)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:58 2006