
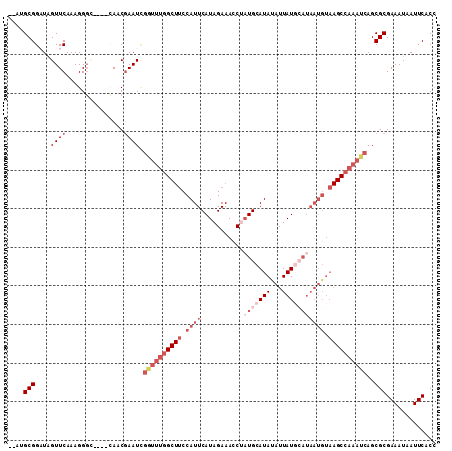
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 9,019,878 – 9,020,043 |
| Length | 165 |
| Max. P | 0.989982 |

| Location | 9,019,878 – 9,019,983 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -12.96 |
| Energy contribution | -16.14 |
| Covariance contribution | 3.18 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9019878 105 + 23771897 --UUGCGGAUAGUUCAAAGGGG----CAACGAAUCGAUUUGGCUUCCAUUCAUAGAAAUCGAUGCAUAUAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACC --(((((....((((....)))----)........((((((((((.((((....(....).(((((((...))))))))))).))))))))))..)))))........... ( -29.10) >DroSec_CAF1 58764 105 + 1 --UUGCGGAUAGUUCAAAGGGG----AAUCGAAUCGGUUUGGCUUCCAUUCAUAGAAACCUAUGCAUAUAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACC --((((((((.((((......)----)))...)))((((((((((.((((.((((....))))(((((...)))))..)))).))))))))))..)))))........... ( -28.50) >DroSim_CAF1 59907 105 + 1 --UUGCGGAUAGUUCAAAGGGC----CAUCGAAUCGGUUUGGCUUCCAUUCAUAGAAACCGAUGCAUAAAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACC --((((((((.(((.....)))----.))).....((((((((((.((((...........(((((((...))))))))))).))))))))))..)))))........... ( -29.30) >DroEre_CAF1 62239 100 + 1 GAAGGCGGUUAGUUCAAAGGGC----CAACCAAUUGGUUUGGCUUCCAUUCAUAGAAACCUAUGCAUAUAUUAU-------GUAAGCCAAAUCAGCGCGAAAUAAUUCACC ....((((((.(((.....)))----.))))..((((((((((((.(((.(((((....)))))........))-------).)))))))))))).))............. ( -27.60) >DroYak_CAF1 61449 107 + 1 AAAAGCGGAUAGUUCAAAGGGC----CAGCGAAUUGGUUUGGCUUCCAUUCAUAGAAACCUAUGCAUAUAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACC ......((((.(((.....)))----..(((...(((((((((((.((((.((((....))))(((((...)))))..)))).))))))))))).)))......))))... ( -28.40) >DroPer_CAF1 58802 96 + 1 CAAGGCGGCUGGUUCAAAGGUUUUCAAAACGAAU----------CCCAUUCAUAGAAAACUUUGCAUAGAUUAUUAAUAAUC-----AAUCCCAGCGCGAAAUAAUUCACU ....(((.((((....(((((((((.....((((----------...))))...))))))))).....(((((....)))))-----....)))))))............. ( -19.50) >consensus __AUGCGGAUAGUUCAAAGGGC____CAACGAAUCGGUUUGGCUUCCAUUCAUAGAAACCUAUGCAUAUAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACC ....(((....((((...............)))).((((((((((.((((...........(((((((...))))))))))).))))))))))..)))............. (-12.96 = -16.14 + 3.18)


| Location | 9,019,911 – 9,020,004 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -14.18 |
| Energy contribution | -16.02 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9019911 93 + 23771897 UGGCUUCCAUUCAUAGAAAUCGAUGCAUAUAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACCAGCCACACGAAGCCGCGUUUC ((((((.((((....(....).(((((((...))))))))))).))))))....((((((((....)))....((........)).))))).. ( -20.20) >DroSec_CAF1 58797 93 + 1 UGGCUUCCAUUCAUAGAAACCUAUGCAUAUAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACCAGCCACACGAAGCCGCGUUUC ((((((.((((.((((....))))(((((...)))))..)))).))))))....((((((((....)))....((........)).))))).. ( -20.60) >DroSim_CAF1 59940 93 + 1 UGGCUUCCAUUCAUAGAAACCGAUGCAUAAAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACCAGCCACACGAAGCAGCGUUUC ((((((.((((...........(((((((...))))))))))).))))))....((((((((....)))....((........)).))))).. ( -20.80) >DroEre_CAF1 62274 86 + 1 UGGCUUCCAUUCAUAGAAACCUAUGCAUAUAUUAU-------GUAAGCCAAAUCAGCGCGAAAUAAUUCACCAGCCACACGAAGUCGCGUUUC ((((((.(((.(((((....)))))........))-------).))))))....(((((((.....(((...........))).))))))).. ( -19.90) >DroYak_CAF1 61484 89 + 1 UGGCUUCCAUUCAUAGAAACCUAUGCAUAUAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACCAGCCACACGAAGCCGCU---- .((((((..............((((((((...)))))))).(((..(((......).))(((....))).......))).))))))...---- ( -18.30) >consensus UGGCUUCCAUUCAUAGAAACCUAUGCAUAUAUUAUGCAUAAUGUAAGCCAAAUCAGCGCGAAAUAAUUCACCAGCCACACGAAGCCGCGUUUC ((((((.((((...........(((((((...))))))))))).))))))....((((((((....)))....((........)).))))).. (-14.18 = -16.02 + 1.84)



| Location | 9,019,911 – 9,020,004 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -21.38 |
| Energy contribution | -22.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9019911 93 - 23771897 GAAACGCGGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCAUAAUAUAUGCAUCGAUUUCUAUGAAUGGAAGCCA .......((((((((((((...(((((....)))))(((.....))))))))).(((((((...)))))))...............)))))). ( -25.00) >DroSec_CAF1 58797 93 - 1 GAAACGCGGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCAUAAUAUAUGCAUAGGUUUCUAUGAAUGGAAGCCA ......(((((......)))))(((((....)))))(((.....)))......((((((((...))))))))((((((((....)))))))). ( -28.50) >DroSim_CAF1 59940 93 - 1 GAAACGCUGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCAUAAUUUAUGCAUCGGUUUCUAUGAAUGGAAGCCA .....((..(.....)..))..(((((....)))))(((.....))).......(((((((...))))))).((((((((....)))))))). ( -26.40) >DroEre_CAF1 62274 86 - 1 GAAACGCGACUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUAC-------AUAAUAUAUGCAUAGGUUUCUAUGAAUGGAAGCCA ...(((((....))))).((..(((((....)))))(((.....)))....-------.........))...((((((((....)))))))). ( -23.10) >DroYak_CAF1 61484 89 - 1 ----AGCGGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCAUAAUAUAUGCAUAGGUUUCUAUGAAUGGAAGCCA ----..(((((......)))))(((((....)))))(((.....)))......((((((((...))))))))((((((((....)))))))). ( -28.50) >consensus GAAACGCGGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCAUAAUAUAUGCAUAGGUUUCUAUGAAUGGAAGCCA .....((.((.....)).))..(((((....)))))(((.....))).......(((((((...))))))).((((((((....)))))))). (-21.38 = -22.22 + 0.84)


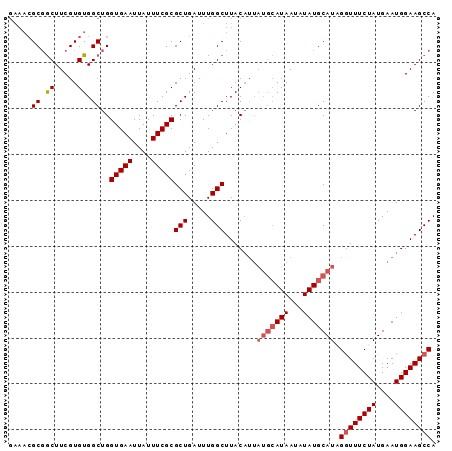
| Location | 9,019,945 – 9,020,043 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -16.97 |
| Energy contribution | -18.58 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 9019945 98 - 23771897 UAAUACAAAACUAAGCCCAAAUUAGUCAAAA-AGCCGCUUGAAACG------------------CGGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCA ...........((((((.((((((((((..(-((((((.......)------------------))))))..))......(((((....)))))))))))))))))))......... ( -31.60) >DroSec_CAF1 58831 98 - 1 UAAUACAAAACUAAGCCCAAAUUAGUCAAAA-AGCCGCUUGAAACG------------------CGGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCA ...........((((((.((((((((((..(-((((((.......)------------------))))))..))......(((((....)))))))))))))))))))......... ( -31.60) >DroSim_CAF1 59974 98 - 1 UAAUACAAAACUAAGCCCAAAUUAGUCAAAA-AGCCGCUUGAAACG------------------CUGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCA ...........((((((.((((((((.....-((((((.((((...------------------....)))).)))))).(((((....)))))))))))))))))))......... ( -30.30) >DroEre_CAF1 62308 92 - 1 UAAUACGAAACUAAGCCGAAAUUAGUCAAAAAAGCAGCUUGAAACG------------------CGACUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUAC-------A ...........((((((.((((((((........(((((....(((------------------((....))))))))))(((((....))))))))))))))))))).-------. ( -30.50) >DroYak_CAF1 61518 88 - 1 UAAUACAAAACUAAGCGCAAAUUAGUCAAAAAA-----------AG------------------CGGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCA ...........(((((.(((((((((.......-----------((------------------(.((.....)).))).(((((....)))))))))))))))))))......... ( -24.80) >DroPer_CAF1 58865 109 - 1 UAAAAUAAAACUAACUUCAAAUUAGUCAA---AGCAACUUUGAACUCGGAGUCGGAAUGGGAUUCGGUUUCGUGUGGCCAGUGAAUUAUUUCGCGCUGGGAUU-----GAUUAUUAA ...................((((((((..---(((........((.((((..((((......))))..)))).)).....(((((....))))))))..))))-----))))..... ( -27.10) >consensus UAAUACAAAACUAAGCCCAAAUUAGUCAAAA_AGCCGCUUGAAACG__________________CGGCUUCGUGUGGCUGGUGAAUUAUUUCGCGCUGAUUUGGCUUACAUUAUGCA ...........((((((.((((((((......................................(((((......)))))(((((....)))))))))))))))))))......... (-16.97 = -18.58 + 1.61)

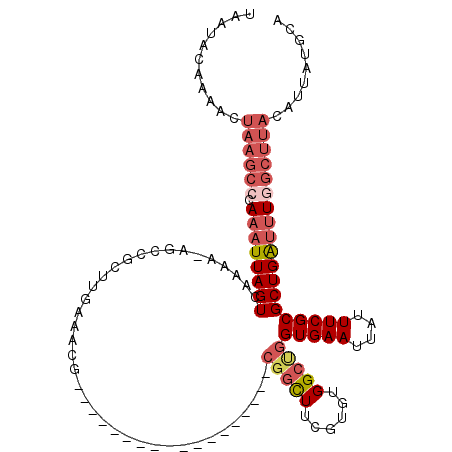

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:49 2006