
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,981,596 – 8,981,700 |
| Length | 104 |
| Max. P | 0.892518 |

| Location | 8,981,596 – 8,981,700 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -38.03 |
| Consensus MFE | -22.75 |
| Energy contribution | -23.45 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
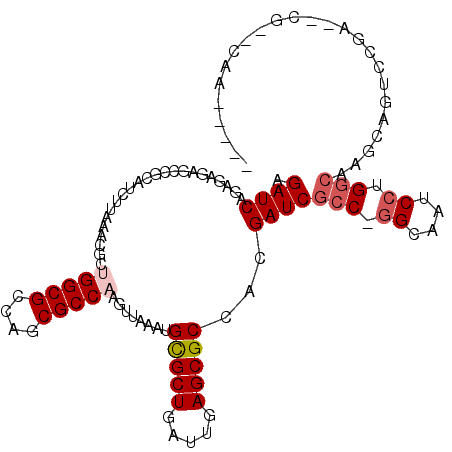
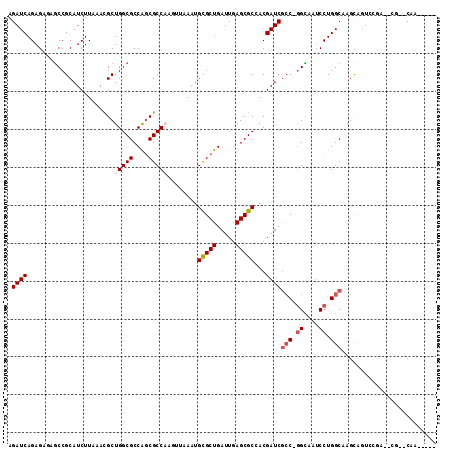
>3L_DroMel_CAF1 8981596 104 + 23771897 AGAUCAGAGCGAGCCGCAUCUUAAACGCUGGCGCCGGCGCCAAGUUAAAUGCGCUGAUUGAGCGCCACGAUCACC-GGCAAACCUGGCGAGCAGUCCAA--UU--CAAA---- ......(((..(((.((((....(((..(((((....))))).)))..)))))))(((((..(((((.(.....)-((....)))))))..)))))...--))--)...---- ( -30.70) >DroVir_CAF1 22987 98 + 1 AGAUCAGAGAGCGCCGCAUCUUAAAUGCUGGCGCCAGCGCC------AGUGCGCUGAUUGAGCGCGACGAUCGCCAGGUAAUCCUGGCCAGCGGUUCGC--CG--CAA----- .((((...(.(((((((((.....)))).)))))).(((((------(((......)))).))))...))))((((((....))))))..((((....)--))--)..----- ( -47.70) >DroGri_CAF1 21943 100 + 1 AGAUCAGAGAGCACCGCAUCUUAAAUGCUGGCGCCAGCGCC------AAUGCGCUGAUUGAGCGCGACGAUCGCCAGGUAAUCCUGGCCAGCCAUGCGC--CAACCAU----- .((((((...(((..((((.....))))(((((....))))------).))).))))))(.(((((......((((((....))))))......)))))--)......----- ( -39.80) >DroWil_CAF1 26140 107 + 1 AGAUCAGAGCAAGCCGCAUCUUAAACGCUGGCGCCA-CGCCAAGUUAAAUGUGCUGAUUGAGCGCCACGAUCGCC-AGCGAUCCUGGCAAGUAGUCGCA--GU--CUCCGGAA ...((.(((...((.((((.((.(((..(((((...-))))).))).)).)))).(((((...((((.(((((..-..))))).))))...))))))).--..--))).)).. ( -35.50) >DroMoj_CAF1 22994 104 + 1 AGAUCAGAGAGAGCCGCAUCUUAAAUGCUGGCGCCAGCGCCAGUGCCAGUGCGCUGAUUGAGCGCGACGAUCGGCAGGUAAUCCUGGCGAGCAGUUGGC--CG--CAA----- ...............((.........(((((((....)))))))(((...(((((.....)))))(((..(((.((((....)))).)))...))))))--.)--)..----- ( -42.00) >DroAna_CAF1 20157 106 + 1 AGAUCAGAGCAAGCCGCAUCUUAAACGCUGGCGCCGGCGCCAAGUUAAAUGCGCUGAUUGAGCGCCACGAUCGCC-GGCAAUCCUGGCAAGCAAUCCGACGUC--CGAC---- .(((((((((.....)).))).....(.((((((((((((..........)))))).....)))))))))))(((-((.....)))))...............--....---- ( -32.50) >consensus AGAUCAGAGAGAGCCGCAUCUUAAACGCUGGCGCCAGCGCCAAGUUAAAUGCGCUGAUUGAGCGCCACGAUCGCC_GGCAAUCCUGGCAAGCAGUCCGA__CG__CAA_____ .((((.......................(((((....)))))........(((((.....)))))...))))(((.((....)).)))......................... (-22.75 = -23.45 + 0.70)

| Location | 8,981,596 – 8,981,700 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -26.61 |
| Energy contribution | -26.42 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
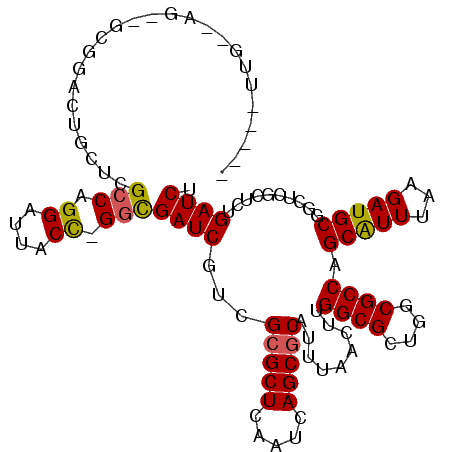
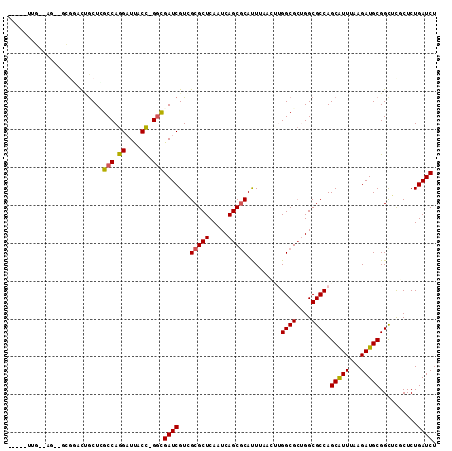
>3L_DroMel_CAF1 8981596 104 - 23771897 ----UUUG--AA--UUGGACUGCUCGCCAGGUUUGCC-GGUGAUCGUGGCGCUCAAUCAGCGCAUUUAACUUGGCGCCGGCGCCAGCGUUUAAGAUGCGGCUCGCUCUGAUCU ----....--.(--(..((..((..(((.(((..(((-((((...((.(((((.....))))).....))....)))))))))).(((((...))))))))..))))..)).. ( -37.50) >DroVir_CAF1 22987 98 - 1 -----UUG--CG--GCGAACCGCUGGCCAGGAUUACCUGGCGAUCGUCGCGCUCAAUCAGCGCACU------GGCGCUGGCGCCAGCAUUUAAGAUGCGGCGCUCUCUGAUCU -----..(--((--((((.....(.((((((....)))))).))))))))(.(((..((((((...------.))))))(((((.(((((...))))))))))....))).). ( -47.30) >DroGri_CAF1 21943 100 - 1 -----AUGGUUG--GCGCAUGGCUGGCCAGGAUUACCUGGCGAUCGUCGCGCUCAAUCAGCGCAUU------GGCGCUGGCGCCAGCAUUUAAGAUGCGGUGCUCUCUGAUCU -----..(((((--((((((((.(.((((((....)))))).))))).)))).)))))(((((...------.)))))((((((.(((((...)))))))))))......... ( -47.60) >DroWil_CAF1 26140 107 - 1 UUCCGGAG--AC--UGCGACUACUUGCCAGGAUCGCU-GGCGAUCGUGGCGCUCAAUCAGCACAUUUAACUUGGCG-UGGCGCCAGCGUUUAAGAUGCGGCUUGCUCUGAUCU ...(((((--..--.((((.....(((((.(((((..-..))))).))))).....)).(((..((.(((((((((-...)))))).))).))..))).))...))))).... ( -39.20) >DroMoj_CAF1 22994 104 - 1 -----UUG--CG--GCCAACUGCUCGCCAGGAUUACCUGCCGAUCGUCGCGCUCAAUCAGCGCACUGGCACUGGCGCUGGCGCCAGCAUUUAAGAUGCGGCUCUCUCUGAUCU -----..(--((--((.....)).)))..((((((...(((((((((((((((.....)))))...))).((((((....)))))).......))).))))......)))))) ( -36.60) >DroAna_CAF1 20157 106 - 1 ----GUCG--GACGUCGGAUUGCUUGCCAGGAUUGCC-GGCGAUCGUGGCGCUCAAUCAGCGCAUUUAACUUGGCGCCGGCGCCAGCGUUUAAGAUGCGGCUUGCUCUGAUCU ----((((--((.((..(((((..(((((.(((((..-..))))).)))))..)))))((((((((((((((((((....)))))).)))..)))))).))).)))))))).. ( -44.80) >consensus _____UUG__AG__GCGGACUGCUCGCCAGGAUUACC_GGCGAUCGUCGCGCUCAAUCAGCGCAUUUAACUUGGCGCUGGCGCCAGCAUUUAAGAUGCGGCUCGCUCUGAUCU .........................(((.((....)).)))((((...(((((.....))))).........((((....)))).(((((...)))))..........)))). (-26.61 = -26.42 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:36 2006