
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,981,333 – 8,981,449 |
| Length | 116 |
| Max. P | 0.993720 |

| Location | 8,981,333 – 8,981,424 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -21.33 |
| Energy contribution | -22.57 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8981333 91 + 23771897 UUCCUCGAGCAAUUUGUGCAUGCUGUAUGCCUUCCAUUUGCCUGUCCUCGCCGAAUACAUAAUUCGGCGUGGUUUUGCCUUUGGUUUCGGU ....(((((........(((((...)))))...(((...((....((.((((((((.....)))))))).))....))...))).))))). ( -25.20) >DroSec_CAF1 20440 88 + 1 UU---CCAGCAAUUUGUGCAUGCUGCAUGCCUUCCUUUUGCCUGUCCUCGCCGAAUACAUAAUUCGGCGUGGUUCUGCCUUUGGUUUCGGU ..---.(((((.........)))))...(((..((....((....((.((((((((.....)))))))).))....))....))....))) ( -25.40) >DroSim_CAF1 21054 91 + 1 UUCCUCCAGCAAUUUGUGCAUGCUGCAUGCCUUCCAUUUGCCUGUCCUCGCCGAAUACAUAAUUCGGCGUGGUUCUGCCUUUGGUUUCGGU ......(((((.........)))))...(((..(((...((....((.((((((((.....)))))))).))....))...)))....))) ( -27.10) >DroEre_CAF1 20852 85 + 1 UU---CCAGCAAUUUGUG--UGCCGCAUGCG-UCCAUUUGCCCGUCCUCGCCGAAUACAUAAUUCGGCUUGGUUUUGCCUUUGGUUUCGGU ..---...((((..((..--(((.....)))-..)).))))(((.((..(((((((.....)))))))..((.....))...))...))). ( -23.50) >consensus UU___CCAGCAAUUUGUGCAUGCUGCAUGCCUUCCAUUUGCCUGUCCUCGCCGAAUACAUAAUUCGGCGUGGUUCUGCCUUUGGUUUCGGU ......(((((.........)))))...(((..(((...((....((.((((((((.....)))))))).))....))...)))....))) (-21.33 = -22.57 + 1.25)

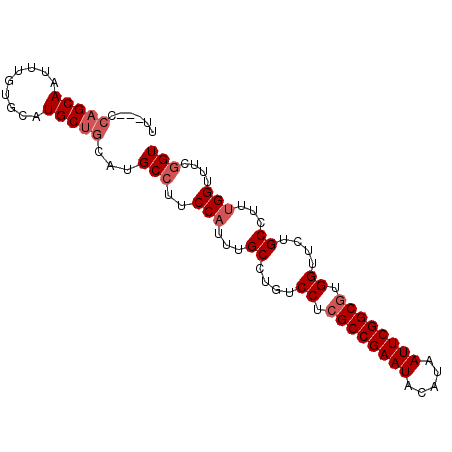

| Location | 8,981,333 – 8,981,424 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.53 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -24.11 |
| Energy contribution | -24.67 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8981333 91 - 23771897 ACCGAAACCAAAGGCAAAACCACGCCGAAUUAUGUAUUCGGCGAGGACAGGCAAAUGGAAGGCAUACAGCAUGCACAAAUUGCUCGAGGAA .((....(((...((....((.((((((((.....)))))))).))....))...)))...((((.....)))).............)).. ( -26.80) >DroSec_CAF1 20440 88 - 1 ACCGAAACCAAAGGCAGAACCACGCCGAAUUAUGUAUUCGGCGAGGACAGGCAAAAGGAAGGCAUGCAGCAUGCACAAAUUGCUGG---AA .(((...((....((....((.((((((((.....)))))))).))....))....))...(((((...))))).........)))---.. ( -26.70) >DroSim_CAF1 21054 91 - 1 ACCGAAACCAAAGGCAGAACCACGCCGAAUUAUGUAUUCGGCGAGGACAGGCAAAUGGAAGGCAUGCAGCAUGCACAAAUUGCUGGAGGAA .((....(((...((....((.((((((((.....)))))))).))....))...)))......(.(((((.........))))).))).. ( -29.80) >DroEre_CAF1 20852 85 - 1 ACCGAAACCAAAGGCAAAACCAAGCCGAAUUAUGUAUUCGGCGAGGACGGGCAAAUGGA-CGCAUGCGGCA--CACAAAUUGCUGG---AA .......(((...((....((..(((((((.....)))))))..))....))...))).-....(.(((((--.......))))).---). ( -24.20) >consensus ACCGAAACCAAAGGCAAAACCACGCCGAAUUAUGUAUUCGGCGAGGACAGGCAAAUGGAAGGCAUGCAGCAUGCACAAAUUGCUGG___AA .......(((...((....((.((((((((.....)))))))).))....))...)))........(((((.........)))))...... (-24.11 = -24.67 + 0.56)

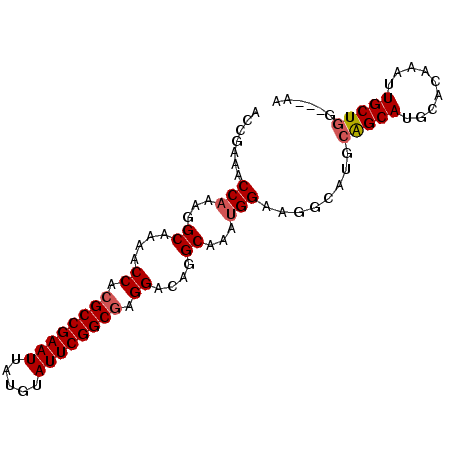

| Location | 8,981,350 – 8,981,449 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -18.40 |
| Energy contribution | -21.36 |
| Covariance contribution | 2.96 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8981350 99 + 23771897 GCAUGCUGUAUGCCUUCCAUUUGCCUGUCCUCGCCGAAUACAUAAUUCGGCGUGGUUUUGCCUUUGGUUUCGGUUUCAUACAUAUCUCCCGCAUGCGAG ((((((((((((((..(((...((....((.((((((((.....)))))))).))....))...)))....))...))))))........))))))... ( -34.80) >DroSec_CAF1 20454 99 + 1 GCAUGCUGCAUGCCUUCCUUUUGCCUGUCCUCGCCGAAUACAUAAUUCGGCGUGGUUCUGCCUUUGGUUUCGGUUUCAUACAUAUCUCCCGCAUGCGAG ((((((((...(((..((....((....((.((((((((.....)))))))).))....))....))....)))..))............))))))... ( -29.40) >DroSim_CAF1 21071 99 + 1 GCAUGCUGCAUGCCUUCCAUUUGCCUGUCCUCGCCGAAUACAUAAUUCGGCGUGGUUCUGCCUUUGGUUUCGGUUUCAUACAAAUCUCCCGCAUGCGAG ((((((..........(((...((....((.((((((((.....)))))))).))....))...)))....(((((.....)))))....))))))... ( -31.70) >DroEre_CAF1 20866 92 + 1 G--UGCCGCAUGCG-UCCAUUUGCCCGUCCUCGCCGAAUACAUAAUUCGGCUUGGUUUUGCCUUUGGUUUCGGUUUCAU----AUCUCCCGCAUGCGAG .--...((((((((-.(((...((....((..(((((((.....)))))))..))....))...)))....(((.....----)))...)))))))).. ( -31.30) >DroYak_CAF1 21518 76 + 1 -------------------CUCACCCUUCCCCACCGCAUACAUAAUUCGGCUUGGUUUUGCCUUUGGUUUCGGUUUCAU----AUCUCCCGCAUGCGCG -------------------...............(((((.........(((........)))........(((......----.....))).))))).. ( -10.80) >consensus GCAUGCUGCAUGCCUUCCAUUUGCCUGUCCUCGCCGAAUACAUAAUUCGGCGUGGUUUUGCCUUUGGUUUCGGUUUCAUACA_AUCUCCCGCAUGCGAG ......(((((((...(((...((....((.((((((((.....)))))))).))....))...)))....((...............))))))))).. (-18.40 = -21.36 + 2.96)

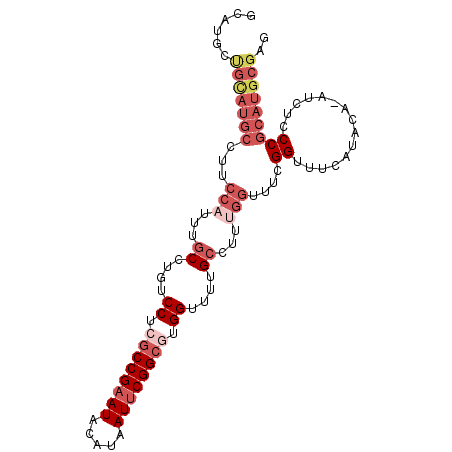
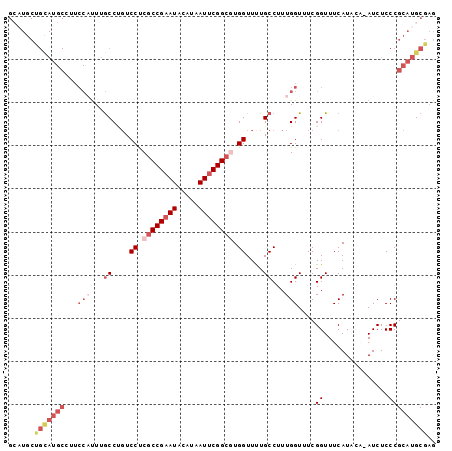
| Location | 8,981,350 – 8,981,449 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -22.06 |
| Energy contribution | -24.54 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8981350 99 - 23771897 CUCGCAUGCGGGAGAUAUGUAUGAAACCGAAACCAAAGGCAAAACCACGCCGAAUUAUGUAUUCGGCGAGGACAGGCAAAUGGAAGGCAUACAGCAUGC ...((((((........((((((...((....(((...((....((.((((((((.....)))))))).))....))...)))..)))))))))))))) ( -34.20) >DroSec_CAF1 20454 99 - 1 CUCGCAUGCGGGAGAUAUGUAUGAAACCGAAACCAAAGGCAGAACCACGCCGAAUUAUGUAUUCGGCGAGGACAGGCAAAAGGAAGGCAUGCAGCAUGC ...((((((........((((((...((....((....((....((.((((((((.....)))))))).))....))....))..)))))))))))))) ( -32.50) >DroSim_CAF1 21071 99 - 1 CUCGCAUGCGGGAGAUUUGUAUGAAACCGAAACCAAAGGCAGAACCACGCCGAAUUAUGUAUUCGGCGAGGACAGGCAAAUGGAAGGCAUGCAGCAUGC ...((((((........((((((...((....(((...((....((.((((((((.....)))))))).))....))...)))..)))))))))))))) ( -34.20) >DroEre_CAF1 20866 92 - 1 CUCGCAUGCGGGAGAU----AUGAAACCGAAACCAAAGGCAAAACCAAGCCGAAUUAUGUAUUCGGCGAGGACGGGCAAAUGGA-CGCAUGCGGCA--C .(((((((((((....----......))....(((...((....((..(((((((.....)))))))..))....))...))).-)))))))))..--. ( -34.60) >DroYak_CAF1 21518 76 - 1 CGCGCAUGCGGGAGAU----AUGAAACCGAAACCAAAGGCAAAACCAAGCCGAAUUAUGUAUGCGGUGGGGAAGGGUGAG------------------- ..(((((((((.....----......)).........(((........))).......)))))))...............------------------- ( -16.70) >consensus CUCGCAUGCGGGAGAU_UGUAUGAAACCGAAACCAAAGGCAAAACCACGCCGAAUUAUGUAUUCGGCGAGGACAGGCAAAUGGAAGGCAUGCAGCAUGC ...((((((((...............))....(((...((....((.((((((((.....)))))))).))....))...)))...))))))....... (-22.06 = -24.54 + 2.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:34 2006