
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,975,694 – 8,975,860 |
| Length | 166 |
| Max. P | 0.937374 |

| Location | 8,975,694 – 8,975,784 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 70.45 |
| Mean single sequence MFE | -24.49 |
| Consensus MFE | -14.24 |
| Energy contribution | -14.88 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8975694 90 + 23771897 GAUUUCACCGAGGAGGAGCGCAAGCUGUUCCUAAAGGUAUGUCAUUCA---UAGUUCA-------AGUCAAAUUGGUGCCAC-----UAACCAGCCGAGUUCAAC (((...(((.(((((.(((....))).)))))...)))..))).....---..(((.(-------(.((...(((((.....-----..)))))..)).)).))) ( -26.70) >DroPse_CAF1 30503 91 + 1 GACUUUACAGAAGAAGAGCGCAAGCUUUUCCUAAAGGUGAGUACCCGAAAGAAGAUCA-------UGUCUAAG----GCCAUCUCCUUA---AUGCAUAUUCUUU .((((.((....(((((((....)))))))......))))))........(((((..(-------(((.((((----(......)))))---..))))..))))) ( -23.00) >DroSec_CAF1 14799 90 + 1 GAUUUCACGGAGGAGGAGCGCAAGCUGUUCCUAAAGGUAAGUCGUUCA---UAGUUCA-------UGUCAAAUUGGUGGCAC-----UAACCAACCGGGUUCAAC ((..((.((((((((.(((....))).)))))...(((.(((.(((..---(((((..-------.....)))))..)))))-----).)))..))))).))... ( -26.90) >DroSim_CAF1 15445 90 + 1 GAUUUCACGGAGGAGGAGCGCAAGCUGUUCCUAAAGGUAAGUCGUUCA---UAGUUCA-------UGUCAAAUUGGUGCCAC-----UAACCAACCGGGUUCAAC ((..((.((((((((.(((....))).)))))...(((.(((.((...---(((((..-------.....)))))..)).))-----).)))..))))).))... ( -27.30) >DroEre_CAF1 15158 86 + 1 GAUUUCACGGAGGAGGAGCGUAAGCUGUUCCUAAAGGUAUGUCGUUUA---UAAUUCA-------AGGCAAAUUGGCGCCAC-----U----AACUGAGCACAAC (((....(..(((((.(((....))).)))))...)....)))(((((---.......-------.(((........)))..-----.----...)))))..... ( -21.84) >DroYak_CAF1 15693 87 + 1 GACUUCACGGAGGAGGAGCGUAAGCUGUUCCUAAAGGUAUGUCGUUCA---UAACUUUGCUACAUACAUAAAU------AAU-----U----AAUUAAGUAAAAC .((((..(..(((((.(((....))).)))))...)((((((.((...---.......)).))))))......------...-----.----....))))..... ( -21.20) >consensus GAUUUCACGGAGGAGGAGCGCAAGCUGUUCCUAAAGGUAAGUCGUUCA___UAGUUCA_______AGUCAAAUUGGUGCCAC_____UA___AACCGAGUUCAAC (((....(..(((((.(((....))).)))))...)....))).............................................................. (-14.24 = -14.88 + 0.64)



| Location | 8,975,751 – 8,975,860 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.58 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -17.45 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8975751 109 - 23771897 ACGAAUCGAGAAUGGUGGCUCCAGGUGGGCGAAACCCAAAAGCGGCGGCUUUGAGCAGCUGGUGACAAACUGAAAA----GUUGAACUCGGCUGGUUA-----GUGGCACCAAUUUGA .(((((.......(((.((((....((((.....))))(((((....))))))))).)))((((....(((((..(----((((....)))))..)))-----))..)))).))))). ( -35.00) >DroPse_CAF1 30563 107 - 1 GCGGAUGGAGAAGGGCGGCUCAAGGUGGGCAAAGCCCAAAAGCGGUGGUUUUGAGCAGCUGGUAACAAACUGUCAA----AAAGAAUAUGCAU---UAAGGAGAUGGC----CUUAGA ............((((.((((.....))))...))))....(((....((((((.(((.((....))..)))))))----))......))).(---(((((......)----))))). ( -28.90) >DroEre_CAF1 15215 105 - 1 ACGAAUCGAGAAGGGUGGCUCCAGGUGGGCGAAACCCAAAAGCGGAGGCUUUGAGCAGCUAGUAACAAACUGGAAA----GUUGUGCUCAGUU----A-----GUGGCGCCAAUUUGC ......((((...((((((((....((((.....))))(((((....)))))))))((((((((...((((....)----))).)))).))))----.-----....))))..)))). ( -29.70) >DroYak_CAF1 15757 99 - 1 ACGAAUCGAGAAGGGUGGCUCCAGGUGGGCGAAACCCAAAAGCGGAGGCUUUGAGCAGCUGGUGACAAACUGAAAA----GUUUUACUUAAUU----A-----AUU------AUUUAU ...(((..((..((((.((.((....))))...)))).......((((((((...(((.((....))..))).)))----))))).))..)))----.-----...------...... ( -24.20) >DroAna_CAF1 14365 100 - 1 GCGAAUGGAGAACGGUGGCUCCAGAUGGGCGAAGCCCAAAAGCGGUGGCUUCGAGCAACUGGUUACAAACUGAAAGAAAAUAUGAA----GUU---UA-----------AGAAUAUGA ............((((....((((.((..((((((((.......).)))))))..)).))))......))))..............----...---..-----------......... ( -23.10) >DroPer_CAF1 16235 107 - 1 GCGGAUGGAGAAGGGCGGCUCAAGGUGGGCAAAGCCCAAAAGCGGUGGUUUUGAGCAGCUGGUAACAAACUGUCAA----AAAGAAUAUGCAU---UAAGGAGAUGGC----CUUAGA ............((((.((((.....))))...))))....(((....((((((.(((.((....))..)))))))----))......))).(---(((((......)----))))). ( -28.90) >consensus ACGAAUCGAGAAGGGUGGCUCCAGGUGGGCGAAACCCAAAAGCGGUGGCUUUGAGCAGCUGGUAACAAACUGAAAA____GAUGAACUUGGUU___UA_____AUGGC____AUUUGA .............(((.((((....((((.....))))(((((....))))))))).)))(((.....)))............................................... (-17.45 = -16.90 + -0.55)

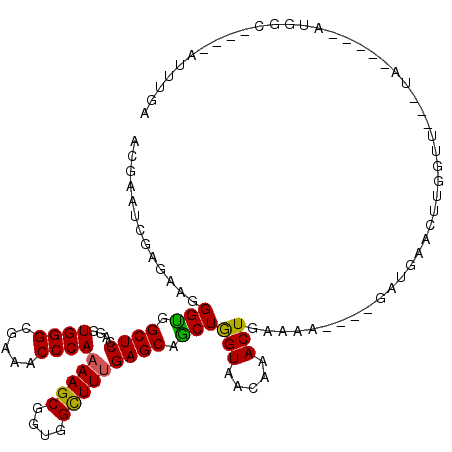
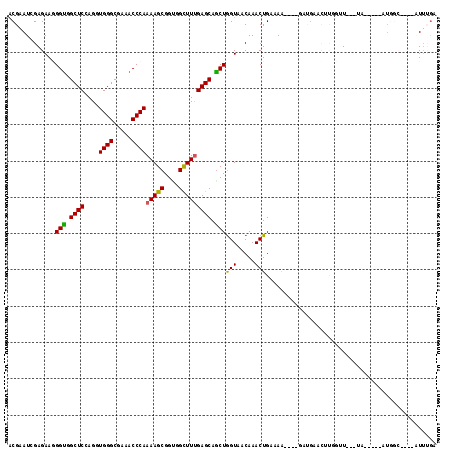
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:30 2006