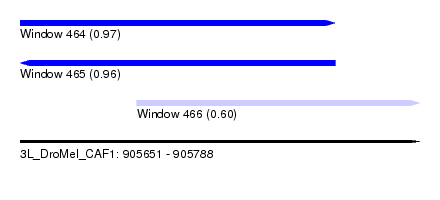
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 905,651 – 905,788 |
| Length | 137 |
| Max. P | 0.965808 |

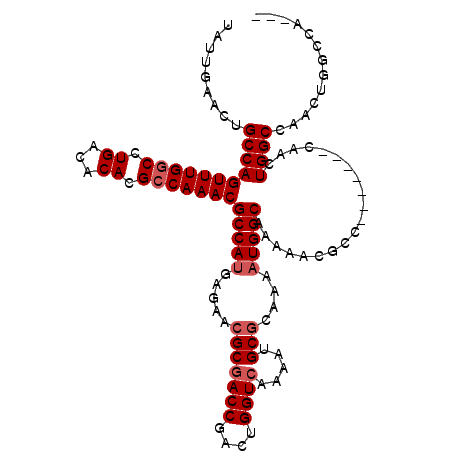
| Location | 905,651 – 905,759 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 87.35 |
| Mean single sequence MFE | -33.31 |
| Consensus MFE | -25.85 |
| Energy contribution | -26.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 905651 108 + 23771897 UAUUGAACUGCCAGUUUGGCCUGACACACGCCAAACGCCAUGAGAACGCGACCGACUGGUCAAAAUGCGCAAAAUGGCAAAAACGCC-------CAACUGGCCAACUGGGCAACU ........((((.(((((((.((...)).)))))))(((((.....(((((((....)))).....)))....)))))......(((-------.....)))......))))... ( -33.50) >DroSec_CAF1 48169 105 + 1 UAUUGAACUGCCAGUUUGGCCUGACACACGACAAACGCCAUGAGAACGCGACCGACUGGUCAAAAUGCGCAAAAUGGCAAAAACGCC-------CAACUGGCCAACUGGGCA--- ..........((((((.((((.......((......(((((.....(((((((....)))).....)))....))))).....))..-------.....))))))))))...--- ( -30.14) >DroEre_CAF1 51479 102 + 1 UAUUGAACUGCCAGUUUGGCCUGACACACGCCAAACGCCAUGAGAACGCUACCGCCUGGUCAAAAUGCGCAAACUGGCAAAAA--------CGCCAACUGGCCAACUGGC----- .........(((((((.((((((((.((.((.....((.........))....)).))))))............((((.....--------.))))...)))))))))))----- ( -30.20) >DroYak_CAF1 50424 115 + 1 UAUUGAACUGCCAGUUUGGCCUGACACACGCCAAACGCCAUGAGAACGCGACCAACUGGUCAAAAUGCACAAAAUGGCAAAAACGCCAACUGGCGAACUGGCGAACUGGCAAACU ........((((((((((......))..(((((..(((((.........((((....)))).............((((......))))..)))))...))))))))))))).... ( -39.40) >consensus UAUUGAACUGCCAGUUUGGCCUGACACACGCCAAACGCCAUGAGAACGCGACCGACUGGUCAAAAUGCGCAAAAUGGCAAAAACGCC_______CAACUGGCCAACUGGCCA___ .........(((((((((((.((...)).)))))))(((((.....(((((((....)))).....)))....)))))....................))))............. (-25.85 = -26.85 + 1.00)

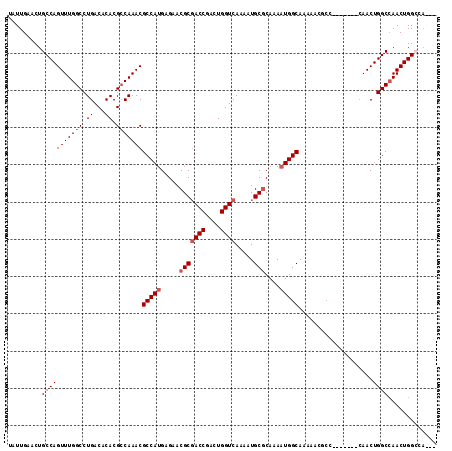
| Location | 905,651 – 905,759 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 87.35 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -31.06 |
| Energy contribution | -32.38 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 905651 108 - 23771897 AGUUGCCCAGUUGGCCAGUUG-------GGCGUUUUUGCCAUUUUGCGCAUUUUGACCAGUCGGUCGCGUUCUCAUGGCGUUUGGCGUGUGUCAGGCCAAACUGGCAGUUCAAUA ..(((((.((((((((...((-------.(((((..((((((...((((.....((((....))))))))....))))))...))))).))...)))).)))))))))....... ( -41.20) >DroSec_CAF1 48169 105 - 1 ---UGCCCAGUUGGCCAGUUG-------GGCGUUUUUGCCAUUUUGCGCAUUUUGACCAGUCGGUCGCGUUCUCAUGGCGUUUGUCGUGUGUCAGGCCAAACUGGCAGUUCAAUA ---((((.((((((((...((-------.(((....((((((...((((.....((((....))))))))....)))))).....))).))...)))).))))))))........ ( -37.70) >DroEre_CAF1 51479 102 - 1 -----GCCAGUUGGCCAGUUGGCG--------UUUUUGCCAGUUUGCGCAUUUUGACCAGGCGGUAGCGUUCUCAUGGCGUUUGGCGUGUGUCAGGCCAAACUGGCAGUUCAAUA -----(((((((((((..((((((--------....))))))...........((((((.((.(.((((((.....))))))).)).)).)))))))).)))))))......... ( -42.50) >DroYak_CAF1 50424 115 - 1 AGUUUGCCAGUUCGCCAGUUCGCCAGUUGGCGUUUUUGCCAUUUUGUGCAUUUUGACCAGUUGGUCGCGUUCUCAUGGCGUUUGGCGUGUGUCAGGCCAAACUGGCAGUUCAAUA ...((((((((((((((....(((((.(((((....)))))..))).)).....((((....)))).........)))))..((((.((...)).)))))))))))))....... ( -43.30) >consensus ___UGCCCAGUUGGCCAGUUG_______GGCGUUUUUGCCAUUUUGCGCAUUUUGACCAGUCGGUCGCGUUCUCAUGGCGUUUGGCGUGUGUCAGGCCAAACUGGCAGUUCAAUA .....(((((((((((.....................(((((...((((.....((((....))))))))....)))))...((((....)))))))).)))))))......... (-31.06 = -32.38 + 1.31)

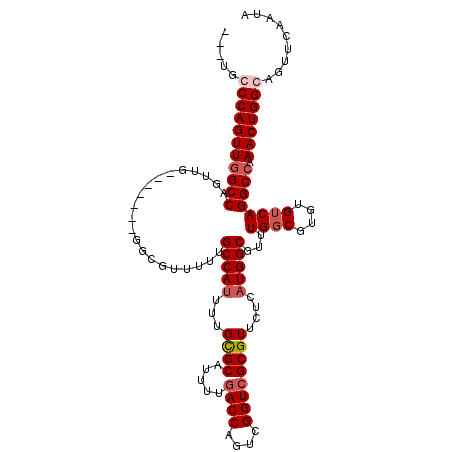
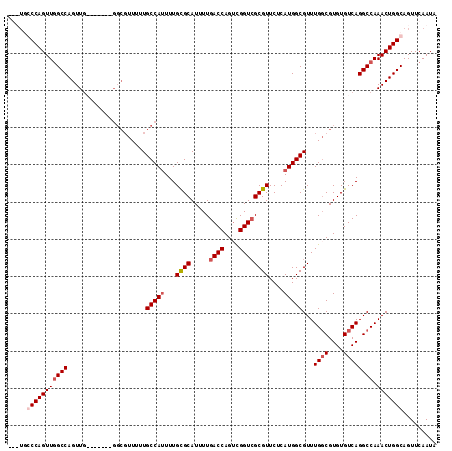
| Location | 905,691 – 905,788 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
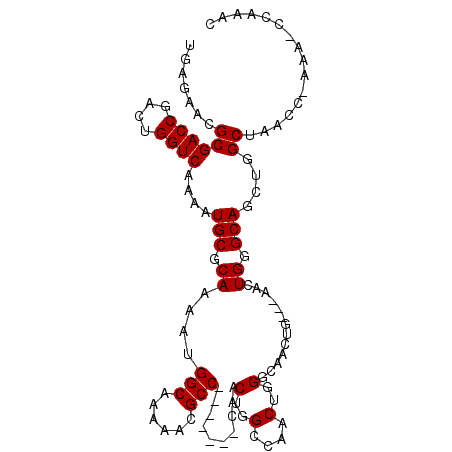
>3L_DroMel_CAF1 905691 97 + 23771897 UGAGAACGCGACCGACUGGUCAAAAUGCGCAAAAUGGCAAAAACGCC-------CAACUGGCCAACUGGGCAACUGGGCAACUGGGCAGCUGGCUAACC------CAAAC .......((((((....))))....(((((......))......(((-------((..((.((....)).))..)))))......)))))(((.....)------))... ( -28.40) >DroSec_CAF1 48209 95 + 1 UGAGAACGCGACCGACUGGUCAAAAUGCGCAAAAUGGCAAAAACGCC-------CAACUGGCCAACUGGGCA--------ACUGGGCAGCUGGCUAACCUAAACCCAAAC .......((((((....)))).....((((......))......(((-------((..((.((....)).))--------..))))).))..))................ ( -26.00) >DroYak_CAF1 50464 105 + 1 UGAGAACGCGACCAACUGGUCAAAAUGCACAAAAUGGCAAAAACGCCAACUGGCGAACUGGCGAACUGGCAAACUGACGAACUGUGCAACUGGC-----GAAAUCGGAAC .((...(((((((....))))....((((((............((((....)))).....((......))............))))))....))-----)...))..... ( -28.00) >consensus UGAGAACGCGACCGACUGGUCAAAAUGCGCAAAAUGGCAAAAACGCC_______CAACUGGCCAACUGGGCAACUG___AACUGGGCAGCUGGCUAACC_AAA_CCAAAC .......((((((....))))....(((.((....(((......)))..........(..(....)..).............)).)))....))................ (-16.80 = -16.80 + 0.00)

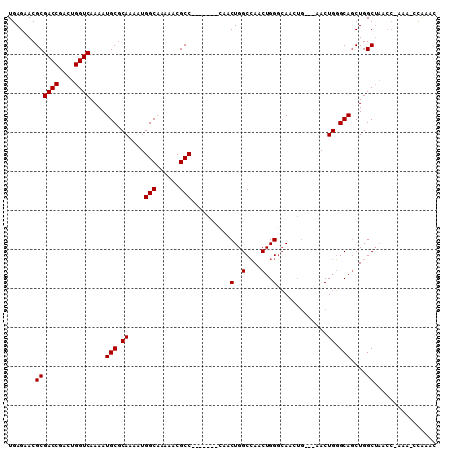
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:57 2006