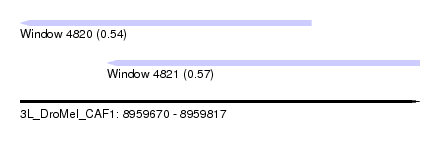
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,959,670 – 8,959,817 |
| Length | 147 |
| Max. P | 0.567244 |

| Location | 8,959,670 – 8,959,777 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -15.60 |
| Energy contribution | -17.79 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
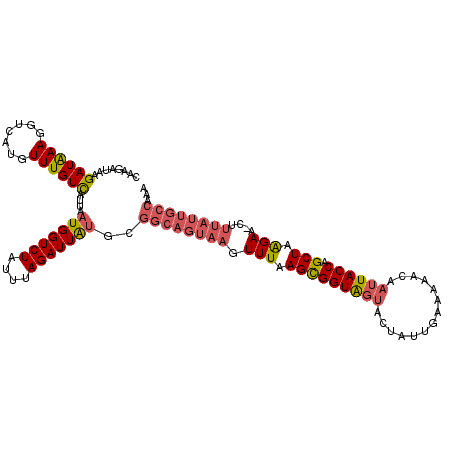
>3L_DroMel_CAF1 8959670 107 - 23771897 UUAGAUUAUGCGGCAGUAAGUUUAAGCGGUAGUACUUUUGAAAAACAAUUACCAGCUAAGAA-CUUUAUUGCCAAAUAAUUUACUAUUAGUUGG--GACUAGACAUAAAC .((((((((..((((((((((((.(((((((((....((....))..)))))).)))..)))-).))))))))..))))))))...(((((...--.)))))........ ( -26.70) >DroSec_CAF1 16934 109 - 1 UUAGAUUAUGCGGCAGUAAGUUUAAGUGGUAGUACUAUUUAAAAACAAUUACCAGCUAAGAA-UUUUAUUGCCAAAUGAUUUACAAUUAGUUGGGGGACUAGACAUGAAC .((((((((..((((((((.(((.(((((((((..............))))))).)).))).-..))))))))..))))))))...(((.((((....))))...))).. ( -26.34) >DroSim_CAF1 16953 109 - 1 UUAGAUUGUGCGGCAGUAAGUUUAAGCGGUAGUACUAUUUAAAAACAAUUACCAGCUAAGAA-UUUUAUUGCCAAAUGAUUUACAAUUUGUUGGGGGAUUAGACAUGAAC .....(..(((((((((((.(((.(((((((((..............)))))).))).))).-..))))))))...((((((.(((....)))..))))))).))..).. ( -23.44) >DroEre_CAF1 17601 97 - 1 UUAGAUUAAG----------UUUAAGCGGUGAUACUUACGAAAUACAUAUACCAGCUUGGAUCCUUUAUCAACAAAUGAUUUGCUAUUAGUUAG--CACUCGACAUGG-U ...(((.(((----------(((((((((((..................)))).)))))))..))).)))......(((..(((((.....)))--)).)))......-. ( -15.37) >consensus UUAGAUUAUGCGGCAGUAAGUUUAAGCGGUAGUACUAUUGAAAAACAAUUACCAGCUAAGAA_CUUUAUUGCCAAAUGAUUUACAAUUAGUUGG__GACUAGACAUGAAC .((((((((..((((((((.(((.(((((((((..............)))))).))).)))....))))))))..))))))))...((((((....))))))........ (-15.60 = -17.79 + 2.19)


| Location | 8,959,702 – 8,959,817 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -16.49 |
| Energy contribution | -18.11 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8959702 115 - 23771897 CGAGAUAAGAUAAAUGUCAUGUUUGUCAUUAAUGGUCUAUUUAGAUUAUGCGGCAGUAAGUUUAAGCGGUAGUACUUUUGAAAAACAAUUACCAGCUAAGAA-CUUUAUUGCCAAA ........(((((((.....)))))))....(((((((....)))))))..((((((((((((.(((((((((....((....))..)))))).)))..)))-).))))))))... ( -29.40) >DroSec_CAF1 16968 115 - 1 CAAGAUAAGAUAAAGGUCAUGUUUGUCAUUAAUGGUCUAUUUAGAUUAUGCGGCAGUAAGUUUAAGUGGUAGUACUAUUUAAAAACAAUUACCAGCUAAGAA-UUUUAUUGCCAAA ........((((((.......))))))....(((((((....)))))))..((((((((.(((.(((((((((..............))))))).)).))).-..))))))))... ( -23.94) >DroSim_CAF1 16987 114 - 1 CAAGAUAAGAUAAAGGUGAUGUUUGUCAU-AAUGGUCUAUUUAGAUUGUGCGGCAGUAAGUUUAAGCGGUAGUACUAUUUAAAAACAAUUACCAGCUAAGAA-UUUUAUUGCCAAA ..(((((.(((....(((((....)))))-....)))))))).........((((((((.(((.(((((((((..............)))))).))).))).-..))))))))... ( -26.54) >DroEre_CAF1 17632 106 - 1 CAAGAUAAGAUGAAAGCCAUGUUUGUUAUUAAUGGUCUAUUUAGAUUAAG----------UUUAAGCGGUGAUACUUACGAAAUACAUAUACCAGCUUGGAUCCUUUAUCAACAAA ........((((((.(((((...........))))).............(----------(((((((((((..................)))).))))))))..))))))...... ( -16.37) >consensus CAAGAUAAGAUAAAGGUCAUGUUUGUCAUUAAUGGUCUAUUUAGAUUAUGCGGCAGUAAGUUUAAGCGGUAGUACUAUUGAAAAACAAUUACCAGCUAAGAA_CUUUAUUGCCAAA ........((((((.......))))))....(((((((....)))))))..((((((((.(((.(((((((((..............)))))).))).)))....))))))))... (-16.49 = -18.11 + 1.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:19 2006