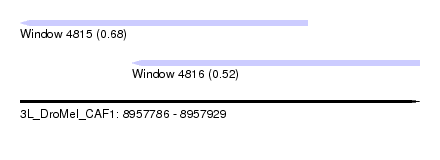
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,957,786 – 8,957,929 |
| Length | 143 |
| Max. P | 0.678052 |

| Location | 8,957,786 – 8,957,889 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 90.58 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -15.09 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
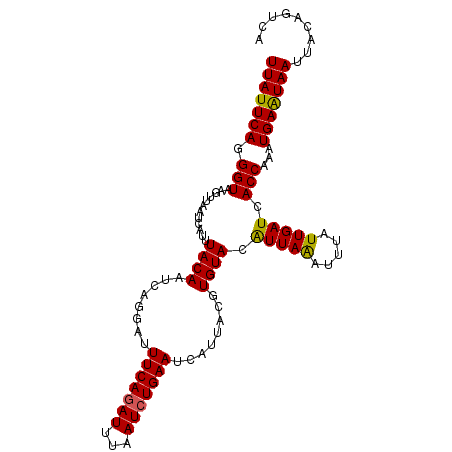
>3L_DroMel_CAF1 8957786 103 - 23771897 UUAUUCAGGGUAAGUUAAUCAUUUACAAUCAGGAUUUCACAUUUAAUCUGAAUCAUUACGUGUACAUUAAAUUUAUUGAUCACCAAAUGAAUAAUUACAGUCA (((((((.(((..((((((.(((((...((((.(((........)))))))........(....)..)))))..)))))).)))...)))))))......... ( -12.40) >DroSec_CAF1 15019 103 - 1 UUAUUCAGGGUAAGUUAAUGAUUUACAAUCAGAAUUUCAGAUUAAAUCUGAAUCAUUACGUGUACAUUAAAUUUAUUGAUCACCAAAUGAAUAAUUACAGUCA (((((((.(((..((((((((..((((........(((((((...)))))))........))))........)))))))).)))...)))))))......... ( -18.89) >DroSim_CAF1 15038 102 - 1 UUAUUCAGGGUAAGUUAAUCAUUUACAAUCAGGAUUUCAGAUUAAAUCUGAAUCA-UACGUGUACAUUAAAUUUAUUGAUCACCAAAUGAAUAAUUACAGUCA (((((((.(((..((((((.(((((((.....((((.(((((...))))))))).-....)))).....)))..)))))).)))...)))))))......... ( -17.40) >DroEre_CAF1 15663 103 - 1 UUAUUCAGGGUAGGUUAAUAAUUUACAAUCAGGAUUUCAGAUUUAAUCUGAAAUAUUGCAUGUACGUUAGGUUUAUUGAUCACCAAAUGAAUAAUUGCACUCA (((((((.(((.(((((((((..((((..(((.(((((((((...))))))))).)))..))))(....)..))))))))))))...)))))))......... ( -26.00) >DroYak_CAF1 15116 103 - 1 UUAUUCAGGGUAGGUUAAUCAUUUACAAUCAGGAUUUCAGAUUUAAUCUGAAAUAUUGCAUGUACGUUAAGUUUAUUGAUAACCAAAUGAGUAAUUACGCUCC (((((((.(((............((((..(((.(((((((((...))))))))).)))..)))).(((((.....))))).)))...)))))))......... ( -20.70) >consensus UUAUUCAGGGUAAGUUAAUCAUUUACAAUCAGGAUUUCAGAUUUAAUCUGAAUCAUUACGUGUACAUUAAAUUUAUUGAUCACCAAAUGAAUAAUUACAGUCA (((((((.(((............((((........(((((((...)))))))........)))).(((((.....))))).)))...)))))))......... (-15.09 = -14.73 + -0.36)

| Location | 8,957,826 – 8,957,929 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -18.35 |
| Energy contribution | -19.77 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
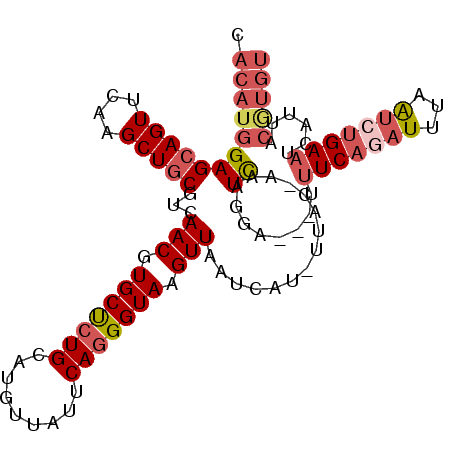
>3L_DroMel_CAF1 8957826 103 - 23771897 CACAUGGAGCAGUUCAAGCUGCGUCAACGUGCUCUGCAUGUUAUUCAGGGUAAGUUAAUCAU-UUAC-AAUCAGGA---UUUCACAUUUAAUCUGAAUCAUUACGUGU .((((((((((((....))))).))(((((((...)))))))((((((..(((((.((((..-....-......))---))....)))))..)))))).....))))) ( -25.20) >DroSec_CAF1 15059 103 - 1 CACAUGGAGCAGUUCAAGCUGCGUCAACGUGCUCUGCAUGUUAUUCAGGGUAAGUUAAUGAU-UUAC-AAUCAGAA---UUUCAGAUUAAAUCUGAAUCAUUACGUGU .((((((((((((....))))).))(((((((...)))))))((((((((((((((...)))-))))-((((.(..---...).))))...))))))).....))))) ( -26.80) >DroSim_CAF1 15078 102 - 1 CACAUGGAGCAGUUCAAGCUGCGUCAACGUGCUCUGCAUGUUAUUCAGGGUAAGUUAAUCAU-UUAC-AAUCAGGA---UUUCAGAUUAAAUCUGAAUCA-UACGUGU .((((((((((((....))))).))(((((((...)))))))(((((((((((((.....))-))))-((((....---.....))))...)))))))..-..))))) ( -25.90) >DroEre_CAF1 15703 103 - 1 CACAUGGAGCAGUUCAAGCUGCGUCAACGUGCUCUGCAUGUUAUUCAGGGUAGGUUAAUAAU-UUAC-AAUCAGGA---UUUCAGAUUUAAUCUGAAAUAUUGCAUGU .((((((((((((....))))).))(((.(((((((.(......)))))))).)))......-....-...(((.(---((((((((...))))))))).)))))))) ( -29.80) >DroYak_CAF1 15156 103 - 1 CACAUGGAGCAGUUCAAGCUGCGUCAACGUGCUCUGCAUGUUAUUCAGGGUAGGUUAAUCAU-UUAC-AAUCAGGA---UUUCAGAUUUAAUCUGAAAUAUUGCAUGU .((((((((((((....))))).))(((.(((((((.(......)))))))).)))......-....-...(((.(---((((((((...))))))))).)))))))) ( -29.80) >DroAna_CAF1 14397 97 - 1 CAUAUGGAGCAGUUCAAGCUCCAACAACGUGCCCUGCAUGUUAUCCAAGGUAGGUUGGUUGUAUUACUGAUUAAGAAACCCUCUCAUUUCGAUUGAU----------- ....((((((.......))))))..(((((((...)))))))...(.((((..(..((((...(((.....)))..))))..)..)))).)......----------- ( -20.70) >consensus CACAUGGAGCAGUUCAAGCUGCGUCAACGUGCUCUGCAUGUUAUUCAGGGUAAGUUAAUCAU_UUAC_AAUCAGGA___UUUCAGAUUUAAUCUGAAUCAUUACGUGU .((((((((((((....)))))...(((.(((((((.........))))))).)))..............))........(((((((...)))))))......))))) (-18.35 = -19.77 + 1.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:14 2006