
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,947,376 – 8,947,474 |
| Length | 98 |
| Max. P | 0.662197 |

| Location | 8,947,376 – 8,947,474 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.39 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -22.23 |
| Energy contribution | -22.45 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
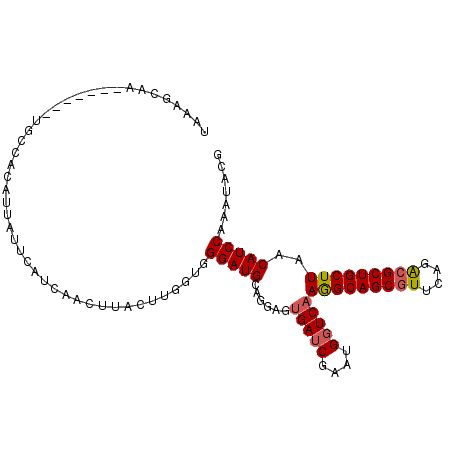
>3L_DroMel_CAF1 8947376 98 + 23771897 UUAAACAA-------UGGCACAUUAUUCAUCAACUUACUUGGUGGGAUGCAGGAGUGAUCGAAUGGUCAAGGCAGCGUUCAGACGCUGCUUAACAUCCAAAUACG ........-------............((((((.....))))))(((((......(((((....)))))(((((((((....)))))))))..)))))....... ( -28.00) >DroPse_CAF1 16739 97 + 1 --AAGCGGAGUUCUUUCCCAGAUC------UUACUUACUUUGUGGGAUGCAGCAGCGAACGGAUGGUCAAAGCAGCAUUCAGGCGCUGCUUAACAUCCAGAUACG --..((((((....))))...(((------((((.......)))))))...)).......(((((....(((((((........)))))))..)))))....... ( -29.70) >DroSec_CAF1 4534 98 + 1 UAAAGCAA-------UGCCACAUUAUUCAUCAACUUACUUGGUGGGAUGCAGGAGUGAUCGAAUGGUCAAGGCAGCGUUCAGACGCUGCUUAACAUCCAAAUACG ....(((.-------(.((((....................)))).)))).(((.(((((....)))))(((((((((....)))))))))....)))....... ( -28.15) >DroSim_CAF1 4583 98 + 1 UAAAGCAA-------UGCCACAUUAUUCAUCAACUUACUUGGUGGGAUGGAGGAGUGAUCGAAUGGUCAAGGCAGCGUUCAGACGCUGCUUAACAUCCAAAUACG ........-------............((((((.....))))))(((((......(((((....)))))(((((((((....)))))))))..)))))....... ( -28.90) >DroEre_CAF1 5104 98 + 1 UUAAGCGC-------UGCCAUAAAACUCUUCAACCUACUUAGUGGGAUGCAGCAGUGAUCGAAUGGUCAAGGCAGCGUUCAGACGCUGCUUAACAUCCAAAUACG ((((((((-------((((.....(((((.((.(((((...))))).)).)).)))((((....))))..))))))((....))...))))))............ ( -29.20) >DroYak_CAF1 4570 98 + 1 UAAAGCAU-------UGCCACAAAAGUGUUCAACUUACUUAGUGGGAUGCAGGAGUGAUCGAAUGGUCAAGGCAGCGUUCAGACGCUGCUUAACAUCCAAAUACG ....((((-------(.((((..(((((.......))))).))))))))).(((.(((((....)))))(((((((((....)))))))))....)))....... ( -34.00) >consensus UAAAGCAA_______UGCCACAUUAUUCAUCAACUUACUUGGUGGGAUGCAGGAGUGAUCGAAUGGUCAAGGCAGCGUUCAGACGCUGCUUAACAUCCAAAUACG ............................................(((((......(((((....)))))(((((((((....)))))))))..)))))....... (-22.23 = -22.45 + 0.22)


| Location | 8,947,376 – 8,947,474 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.39 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -20.35 |
| Energy contribution | -20.27 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8947376 98 - 23771897 CGUAUUUGGAUGUUAAGCAGCGUCUGAACGCUGCCUUGACCAUUCGAUCACUCCUGCAUCCCACCAAGUAAGUUGAUGAAUAAUGUGCCA-------UUGUUUAA ..(((((((..((((((((((((....)))))).)))))).....(((((....)).)))...)))))))......(((((((((...))-------))))))). ( -26.30) >DroPse_CAF1 16739 97 - 1 CGUAUCUGGAUGUUAAGCAGCGCCUGAAUGCUGCUUUGACCAUCCGUUCGCUGCUGCAUCCCACAAAGUAAGUAA------GAUCUGGGAAAGAACUCCGCUU-- .......(((((..((((((((......))))))))....)))))((((((....)).(((((............------....)))))..)))).......-- ( -28.69) >DroSec_CAF1 4534 98 - 1 CGUAUUUGGAUGUUAAGCAGCGUCUGAACGCUGCCUUGACCAUUCGAUCACUCCUGCAUCCCACCAAGUAAGUUGAUGAAUAAUGUGGCA-------UUGCUUUA .((..((((((((((((((((((....)))))).))))).)))))))..))....(((..((((....................))))..-------.))).... ( -26.35) >DroSim_CAF1 4583 98 - 1 CGUAUUUGGAUGUUAAGCAGCGUCUGAACGCUGCCUUGACCAUUCGAUCACUCCUCCAUCCCACCAAGUAAGUUGAUGAAUAAUGUGGCA-------UUGCUUUA .((..((((((((((((((((((....)))))).))))).)))))))..))..............((((((((((((.....)).)))).-------)))))).. ( -23.50) >DroEre_CAF1 5104 98 - 1 CGUAUUUGGAUGUUAAGCAGCGUCUGAACGCUGCCUUGACCAUUCGAUCACUGCUGCAUCCCACUAAGUAGGUUGAAGAGUUUUAUGGCA-------GCGCUUAA ....((..((((((....))))))..))(((((((..(((..((((((..(((((...........)))))))))))..)))....))))-------)))..... ( -32.40) >DroYak_CAF1 4570 98 - 1 CGUAUUUGGAUGUUAAGCAGCGUCUGAACGCUGCCUUGACCAUUCGAUCACUCCUGCAUCCCACUAAGUAAGUUGAACACUUUUGUGGCA-------AUGCUUUA .((..((((((((((((((((((....)))))).))))).)))))))..))....((((.((((.((((.........))))..))))..-------)))).... ( -30.30) >consensus CGUAUUUGGAUGUUAAGCAGCGUCUGAACGCUGCCUUGACCAUUCGAUCACUCCUGCAUCCCACCAAGUAAGUUGAUGAAUAAUGUGGCA_______UUGCUUUA .((..((((((((((((((((((....))))))).)))).)))))))..))...................................................... (-20.35 = -20.27 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:11 2006