| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,901,924 – 8,902,025 |
| Length | 101 |
| Max. P | 0.944825 |

| Location | 8,901,924 – 8,902,025 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -30.02 |
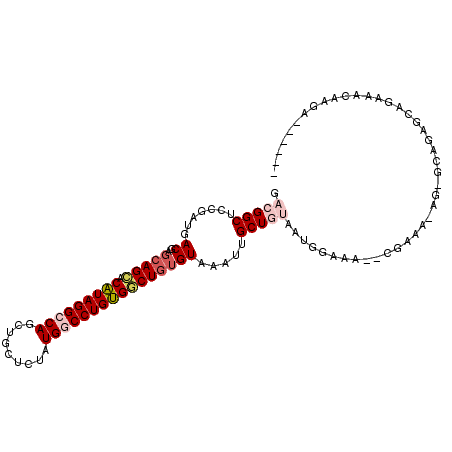
| Consensus MFE | -22.05 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8901924 101 - 23771897 GACGGCUCCGAUGACGAGGAGCACAUAGGCCAGCUGCUCUAUGGCCUGUGGCUGUGUAAAUUGCUGUAAUGGAAA--CGAAAAAGAGCAGAGCAGAAACACGA------ ..(((((((........))))).(((((((((.........))))))))).((((.....(((((.(..((....--))....).))))).)))).....)).------ ( -34.60) >DroGri_CAF1 165389 101 - 1 GACGGCUCCGAUGACGAGCAGAGCGUAGGCCAUUUGCUCUAUGACCUGCGGCUGUGUAAAAUGCUACAA----AA--CGAGAUAA-AC-AAGCAGAGACCAGAAUAAUG ...(((((.........(((.((((((((.(((.......))).))))).))).)))....((((....----..--........-..-.))))))).))......... ( -23.63) >DroSec_CAF1 161654 101 - 1 GACGGCUCCGAUGACGAGCAGCACAUAGGCCAGCUGCUCUAUGGCCUGUGGCUGUGUAAAUUGCUGUAUUGGAAA--CGAAAGAGAGCAGAGCAGAAACAAUA------ ....((((.........(((((.(((((((((.........))))))))))))))......((((.(.(((....--)))...).))))))))..........------ ( -37.50) >DroSim_CAF1 160494 101 - 1 GACGGCUCCGAUGACGAGCAGCACAUAGGCCAGCUGCUCUAUGGCCUGUGGCUGUGUAAAUUGCUGUAAUGGAAA--CGAAAAAGAGCAGAGCAGAAACAAGA------ ....((((.........(((((.(((((((((.........))))))))))))))......((((.(..((....--))....).))))))))..........------ ( -34.80) >DroYak_CAF1 164993 96 - 1 GACGGCUCCGAUGACGAGCAGCACAUAGGCCAGCUGCUCUAUGGCCUGUGGCUGUGUAAAUUGCUGUAAUGGAAA--CGAA-----GCAAAAAAAAAAAUAAA------ ..((....)).......(((((.(((((((((.........)))))))))))))).....(((((....((....--)).)-----)))).............------ ( -31.50) >DroPer_CAF1 187857 92 - 1 GACGGCUCCGAUGACGAUCAGAACGUAGGCCAGCUGCUCUAUGACCUGCGUCUGUGUAAAUUGCUACAAAAGAUAUACGAA-----A------CGAGAAAACA------ ..((..((....((((..(((..(((((((.....)).)))))..))))))).(((((..((.......))..))))))).-----.------))........------ ( -18.10) >consensus GACGGCUCCGAUGACGAGCAGCACAUAGGCCAGCUGCUCUAUGGCCUGUGGCUGUGUAAAUUGCUGUAAUGGAAA__CGAAA_AG_GCAGAGCAGAAACAAGA______ .(((((.......((..(((((.(((((((((.........)))))))))))))))).....))))).......................................... (-22.05 = -22.88 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:57 2006