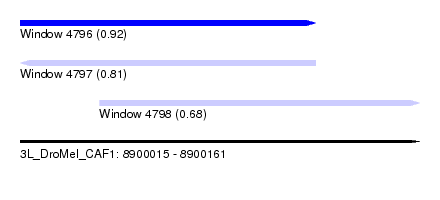
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,900,015 – 8,900,161 |
| Length | 146 |
| Max. P | 0.922067 |

| Location | 8,900,015 – 8,900,123 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
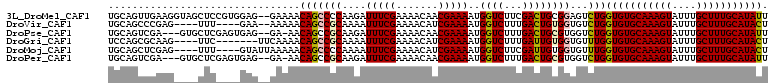
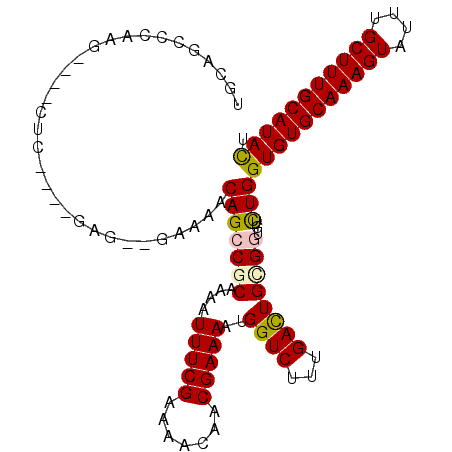
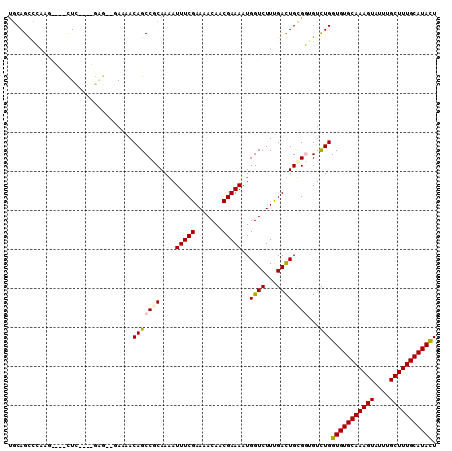
>3L_DroMel_CAF1 8900015 108 + 23771897 UGCAGUUGAAGGUAGCUCCGUGGAG--GAAAACAGCCCCAAGAUUUCGAAAACAACGAAAAUGGUCUUCGACUGCGGAGUCUGGUGUGCAAAGUAUUUGCUUUGCAUAUU .(((((((((((.....((((((.(--(.......))))....(((((.......)))))))))))))))))))).......((((((((((((....)))))))))))) ( -37.10) >DroVir_CAF1 190391 100 + 1 UGCAGCCCGAG----UUU----GAA--AAAAACAGCCGCAAAAUUUCGAAAACAUCGAAAAUGGUCUUUGACUGUGGUGUCUGGUGUGCAAAGUAUUUGCUUUGCAUACU ..(((.....(----(((----...--..)))).(((((....((((((.....))))))..((((...)))))))))..)))(((((((((((....))))))))))). ( -31.00) >DroPse_CAF1 193011 104 + 1 UGCAGUCGA---GUGCUCGAGUGAG--GA-AACAGCCGCAAGAUUUCGAAAACAACGAAAAUGGUCUUUGACUGCGUGGUCUGGUGUGCAAAGUAUUUGCUUUGCAUAUU .((((((((---(.(((((.((..(--..-..).)))).....(((((.......)))))..))).))))))))).......((((((((((((....)))))))))))) ( -33.50) >DroGri_CAF1 162933 99 + 1 UCCAGCGCAAG----UUC-------UUCAAAACAGCCGCAAAAUUUCGAAAACAUCGAAAAUGGUCUUUGAUUGUGGUGUUUGGUGUGCAAAGUAUUUGCUUUGCAUACU ...(((....)----)).-------..((((...((((((...((((((.....))))))...(((...))))))))).))))(((((((((((....))))))))))). ( -30.80) >DroMoj_CAF1 190670 102 + 1 UGCAGCUCGAG----UUU----GUAUUAAAAACAGCCCCAAAAUUUCGAAAACAUCGAAAAUGGUCUUCGAUUGUGGUGUUUGGUGUGCAAAGUAUUUGCUUUGCAUACU .((((.(((((----(((----.......))))....(((...((((((.....)))))).)))...)))))))).......((((((((((((....)))))))))))) ( -28.90) >DroPer_CAF1 185881 104 + 1 UGCAGUCGA---GUGCUCGAGUGAG--GA-AACAGCCGCAAGAUUUCGAAAACAACGAAAAUGGUCUUUGACUGCGUGGUCUGGUGUGCAAAGUAUUUGCUUUGCAUAUU .((((((((---(.(((((.((..(--..-..).)))).....(((((.......)))))..))).))))))))).......((((((((((((....)))))))))))) ( -33.50) >consensus UGCAGCCCAAG____CUC____GAG__GAAAACAGCCGCAAAAUUUCGAAAACAACGAAAAUGGUCUUUGACUGCGGUGUCUGGUGUGCAAAGUAUUUGCUUUGCAUACU ................................(((((((....(((((.......)))))..((((...))))))))...)))(((((((((((....))))))))))). (-21.84 = -21.70 + -0.14)
| Location | 8,900,015 – 8,900,123 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -16.65 |
| Energy contribution | -16.48 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8900015 108 - 23771897 AAUAUGCAAAGCAAAUACUUUGCACACCAGACUCCGCAGUCGAAGACCAUUUUCGUUGUUUUCGAAAUCUUGGGGCUGUUUUC--CUCCACGGAGCUACCUUCAACUGCA ....(((((((......)))))))...........(((((.((((.(((.(((((.......)))))...)))(((.....((--(.....))))))..)))).))))). ( -30.20) >DroVir_CAF1 190391 100 - 1 AGUAUGCAAAGCAAAUACUUUGCACACCAGACACCACAGUCAAAGACCAUUUUCGAUGUUUUCGAAAUUUUGCGGCUGUUUUU--UUC----AAA----CUCGGGCUGCA .((.(((((((......))))))).))..(((......))).........((((((.....))))))...(((((((((((..--...----)))----)...))))))) ( -25.10) >DroPse_CAF1 193011 104 - 1 AAUAUGCAAAGCAAAUACUUUGCACACCAGACCACGCAGUCAAAGACCAUUUUCGUUGUUUUCGAAAUCUUGCGGCUGUU-UC--CUCACUCGAGCAC---UCGACUGCA ....(((((((......)))))))...........((((((.((((....(((((.......)))))))))..(..((((-(.--.......))))).---.))))))). ( -25.50) >DroGri_CAF1 162933 99 - 1 AGUAUGCAAAGCAAAUACUUUGCACACCAAACACCACAAUCAAAGACCAUUUUCGAUGUUUUCGAAAUUUUGCGGCUGUUUUGAA-------GAA----CUUGCGCUGGA .((.(((((((......))))))).))...................(((.((((((.....))))))....(((...((((....-------)))----)...)))))). ( -21.20) >DroMoj_CAF1 190670 102 - 1 AGUAUGCAAAGCAAAUACUUUGCACACCAAACACCACAAUCGAAGACCAUUUUCGAUGUUUUCGAAAUUUUGGGGCUGUUUUUAAUAC----AAA----CUCGAGCUGCA .((.(((((((......))))))).))............((((((((.(((...)))))))))))...((((((..(((.......))----)..----))))))..... ( -22.30) >DroPer_CAF1 185881 104 - 1 AAUAUGCAAAGCAAAUACUUUGCACACCAGACCACGCAGUCAAAGACCAUUUUCGUUGUUUUCGAAAUCUUGCGGCUGUU-UC--CUCACUCGAGCAC---UCGACUGCA ....(((((((......)))))))...........((((((.((((....(((((.......)))))))))..(..((((-(.--.......))))).---.))))))). ( -25.50) >consensus AAUAUGCAAAGCAAAUACUUUGCACACCAGACACCACAGUCAAAGACCAUUUUCGAUGUUUUCGAAAUCUUGCGGCUGUUUUC__CUC____GAA____CUUCGACUGCA ....(((((((......)))))))...........((((((.((((....(((((.......)))))))))..))))))............................... (-16.65 = -16.48 + -0.17)


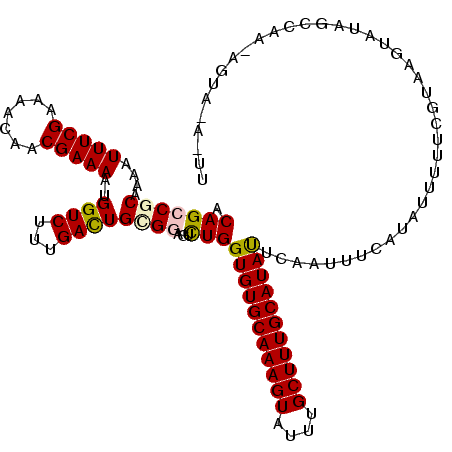
| Location | 8,900,044 – 8,900,161 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -21.88 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8900044 117 + 23771897 ACAGCCCCAAGAUUUCGAAAACAACGAAAAUGGUCUUCGACUGCGGAGUCUGGUGUGCAAAGUAUUUGCUUUGCAUAUUCAAUUUUAUAUUUUUCGUAAGUACAGAUGA-AGUAAU-GU .......((..((((((...((.(((((((.((.(((((....))))).))((((((((((((....))))))))))))...........)))))))..)).....)))-)))..)-). ( -26.90) >DroVir_CAF1 190412 118 + 1 ACAGCCGCAAAAUUUCGAAAACAUCGAAAAUGGUCUUUGACUGUGGUGUCUGGUGUGCAAAGUAUUUGCUUUGCAUACUGAAUUUCAUAUUUUUUGUGAGUAUAA-AUAAAGUACAAUU ...(((((....((((((.....))))))..((((...))))))))).((.((((((((((((....))))))))))))))...(((((.....)))))((((..-.....)))).... ( -33.40) >DroPse_CAF1 193036 110 + 1 ACAGCCGCAAGAUUUCGAAAACAACGAAAAUGGUCUUUGACUGCGUGGUCUGGUGUGCAAAGUAUUUGCUUUGCAUAUUCAAUUUCAUAUUUUUCGUAAGUAUUGCCAA--------U- ......((((.((((((((((....((((.(((.....((((....))))..(((((((((((....)))))))))))))).))))....)))))).)))).))))...--------.- ( -29.30) >DroGri_CAF1 162953 119 + 1 ACAGCCGCAAAAUUUCGAAAACAUCGAAAAUGGUCUUUGAUUGUGGUGUUUGGUGUGCAAAGUAUUUGCUUUGCAUACUAAAUUUCAUAUUCUUUGUAAGUAUACACAAUAUAACAAUU ......(((((.((((((.....)))))).........((.(((((.((((((((((((((((....)))))))))))))))).))))).)))))))...................... ( -33.50) >DroAna_CAF1 157752 117 + 1 ACAGCCCCAAAAUUUCGAAAACAACGAAAAUGGUCUUCGACUGCGGAGUCUGGUGUGCAAAGUAUUUGCUUUGCAUAUUCAAUUUUAUAUUUUUUGUAAGUAUCUCGAG-CGUUAA-GU .............(((((..((.(((((((.((.(((((....))))).))((((((((((((....))))))))))))...........)))))))..))...)))))-......-.. ( -25.80) >DroPer_CAF1 185906 111 + 1 ACAGCCGCAAGAUUUCGAAAACAACGAAAAUGGUCUUUGACUGCGUGGUCUGGUGUGCAAAGUAUUUGCUUUGCAUAUUCAAUUUCAUAUUUUUCGUAAGUAUUGCCAA--------UU ......((((.((((((((((....((((.(((.....((((....))))..(((((((((((....)))))))))))))).))))....)))))).)))).))))...--------.. ( -29.30) >consensus ACAGCCGCAAAAUUUCGAAAACAACGAAAAUGGUCUUUGACUGCGGAGUCUGGUGUGCAAAGUAUUUGCUUUGCAUAUUCAAUUUCAUAUUUUUCGUAAGUAUAGCCAA_AGUA_A_UU .(((((((....(((((.......)))))..((((...))))))))...)))(((((((((((....)))))))))))......................................... (-21.88 = -21.93 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:56 2006