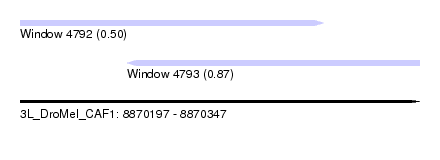
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,870,197 – 8,870,347 |
| Length | 150 |
| Max. P | 0.866732 |

| Location | 8,870,197 – 8,870,311 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.53 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8870197 114 + 23771897 GGUGAAAGUACUUGCCGUAGGAGUUCCACUUCAUCUGCAGAUACUUCUGACUGAGCGGAUGGGCCAGCAGCUCCACCCUGCCAUGUGUUACCAUGGUCUGAUCG--AGGGAUUAU----A ((..(......)..))...(((((((...((((((((((((....)))......)))))))))...).)))))).((((((((((......)))))).......--)))).....----. ( -33.71) >DroVir_CAF1 151580 110 + 1 GAUGAAAAUAUUUGCCAUAAGAGUUCCAUUUCAUUUGCAAAUACUUUUGACUGAGCGGAUGGGCCAGCAGCUCCACGCGUCCAUGCGUGACCAUGGUCUAGUUG--AACAG--------A ...((((.(((((((.....(((......)))....))))))).))))..(((..(((.(((((((.......((((((....))))))....))))))).)))--..)))--------. ( -28.20) >DroGri_CAF1 132444 114 + 1 GAUGAAAGUAUUUGCCAUAAGAGUUCCAUUUCAUUUGCAGAUACUUCUGGCUCAAGGGAUGGGCCAGCAGCUCCACGCGUCCAUGUGUGACCAUUGUCUAGUCA--CAGGAGUAU----A .....((((((((((.....(((......)))....))))))))))((((((((.....))))))))..(((((..(((....)))(((((.........))))--).)))))..----. ( -34.90) >DroWil_CAF1 172723 107 + 1 AAUGAAAGUAUUUGCCAUAUGAAUUCCAUUUCAUUUGCAAGUACUUUUGGCUAAGUGGAUGGGCCAAUAGCUCAACACGUCCAUGGGUGACCAUGGUCUAG--G--G-CGA--------A ...((((((((((((...(((((......)))))..)))))))))))).(((..(((..(((((.....))))).)))(.((((((....)))))).)...--)--)-)..--------. ( -43.40) >DroMoj_CAF1 155111 118 + 1 GAUGAAAGUAUUUGCCAUACGAAUUCCAUUUCAUUUGUAAAUAUUUCUGGCUCAGCGGAUGUGCCAGCAGCUCCACACGUCCAUGGGUGACCAUGGUCUAGGCA--GACGGGAAGUGGGG .....((((((((((.....(((......)))....))))))))))(((((.((.....)).)))))...((((((.(((((((((....)))))((....)).--))))....)))))) ( -38.80) >DroAna_CAF1 128092 116 + 1 GAUGAAAGUAUUUGCCGUAGGAGUUCCACUUCAUCUGUAGAUACUUCUGGCUGAGAGGAUGGGCCAGCAGCUCCACCCGGCCAUGGGUUACCAUAGUCUAUCCCCCAGAAAUCAA----A (((....((((((((.(..((((.....))))..).))))))))((((((..((..((.(((((.....)).))).))(((.((((....)))).)))..))..)))))))))..----. ( -35.70) >consensus GAUGAAAGUAUUUGCCAUAAGAGUUCCAUUUCAUUUGCAAAUACUUCUGGCUGAGCGGAUGGGCCAGCAGCUCCACACGUCCAUGGGUGACCAUGGUCUAGUCG__AACGA__A_____A .....((((((((((.(...(((......)))..).))))))))))((((((.(.....).))))))...........(.(((((......))))).)...................... (-18.82 = -18.43 + -0.38)


| Location | 8,870,237 – 8,870,347 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.72 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -15.99 |
| Energy contribution | -15.43 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8870237 110 - 23771897 AGACUUUAUGCUACCCAUUUCUGAUAUAU--GUUUCCC-UAUAAUCCCUCGAUCAGACCAUGGUAACACAUGGCAGGGUGGAGCUGCUGGCCCAUCCGCUCAGUCAGAAGUAU ..((((....((((((...((((((....--(((....-...)))......))))))(((((......)))))..))))))..((((((((......)).))).))))))).. ( -29.60) >DroPse_CAF1 159096 100 - 1 CAGAUGCUU--------UUCCUAACACA----UUCCCUCUCUGAUCC-CCGGCCAGACCAUGGUUACCCAUGGACGCGUGGAGCUGCUGGCCCAUCCACUCAGCCAGAAGUAU ...((((((--------((.........----........((((...-..((((((.((((((....))))))..((.....))..)))))).......))))..)))))))) ( -26.01) >DroGri_CAF1 132484 108 - 1 CAAAUAAACUAU-----UCACUAAUAUAUAUCUUCUAUAUAUACUCCUGUGACUAGACAAUGGUCACACAUGGACGCGUGGAGCUGCUGGCCCAUCCCUUGAGCCAGAAGUAU ...........(-----((((..(((((((.....)))))))..(((((((((((.....))))))))...)))...)))))(((.(((((.((.....)).))))).))).. ( -30.90) >DroSim_CAF1 127943 112 - 1 CAAAUAUAUCUUUCAAAUUACUAAUAUAUACAUUCCCU-UUUAAUCAAUCGAUCAGACCAUGGUAACCCAUGGCAGGGUGGAGCUGCUGGCCCACCCGCUCAGCCAGAAGUAU ...........................((((.(((...-................(.((((((....))))))).((((((.((.....)))))))).........))))))) ( -26.30) >DroYak_CAF1 133578 110 - 1 CUAUUGCAUUCUUCGCAUUUCUGAUACAU--GUUAUCC-UGUAAUCCCUCGACCAGACCAUGGUUACCCAUGGCAGGGUGGAGCUGCUGGCCCAUCCGCUCAGCCAGAAGUAU .....((.......))(((((((.((((.--(....).-))))....(((.(((...((((((....))))))...))).)))..((((((......)).))))))))))).. ( -33.00) >DroPer_CAF1 151007 100 - 1 CAGAUGCUU--------UUCCUAACACA----UUCCCUCUCUGAUCC-CCGGCCAGACCAUGGUUACCCAUGGACGCGUGGAGCUGCUGGCCCAUCCACUCAGCCAGAAGUAU ...((((((--------((.........----........((((...-..((((((.((((((....))))))..((.....))..)))))).......))))..)))))))) ( -26.01) >consensus CAAAUGCAU__U_____UUACUAAUACAU___UUCCCU_UAUAAUCC_UCGACCAGACCAUGGUUACCCAUGGAAGCGUGGAGCUGCUGGCCCAUCCGCUCAGCCAGAAGUAU .................................................(.((....(((((......)))))....)).).(((.(((((...........))))).))).. (-15.99 = -15.43 + -0.55)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:51 2006