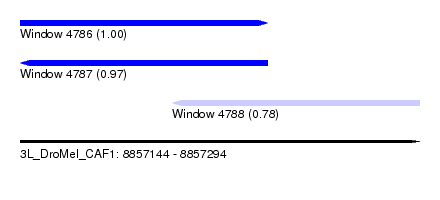
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,857,144 – 8,857,294 |
| Length | 150 |
| Max. P | 0.999185 |

| Location | 8,857,144 – 8,857,237 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.02 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.999185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8857144 93 + 23771897 ------CCUUUACACAGUAGAAGUUGAUUUUCUC-----------------UUCUGAAAAUCUACAUAACUCAAUAUUUGAGUAGCAGGUUCGUUUACUUUUGCCCUGUGUAAAUC ------..((((((((((((((((.(((((((..-----------------....)))))))......(((((.....))))).............))))))))..)))))))).. ( -24.80) >DroSec_CAF1 117047 110 + 1 AAGAACUGCUUACACAGUAG------AUUUUCUCUUUACCCAACUAACUUCUUUUGAAAAUCUACAUAACUCAAUAUUUAAGUAGCAAGUUCUUUUACUUUUGCCCUGUGUAAAUC .........(((((((((((------((((((.......................))))))))))................((((.((((......))))))))..)))))))... ( -20.90) >DroSim_CAF1 113475 116 + 1 AAGUACUGCUUACACAGUAGAAGUUGAUUUUCUCUUUACCCAACUAACUUCUUUUGAAAAUCUACAUAACUCAAUAUUUAAGUAGCAAGUUCUUUUACUUUUGCCCUGUGUAAAUC .........((((((((.....((.(((((((.......................))))))).))................((((.((((......)))))))).))))))))... ( -19.00) >consensus AAG_ACUGCUUACACAGUAGAAGUUGAUUUUCUCUUUACCCAACUAACUUCUUUUGAAAAUCUACAUAACUCAAUAUUUAAGUAGCAAGUUCUUUUACUUUUGCCCUGUGUAAAUC .........((((((((........(((((((.......................)))))))...................((((.((((......)))))))).))))))))... (-15.65 = -15.77 + 0.11)


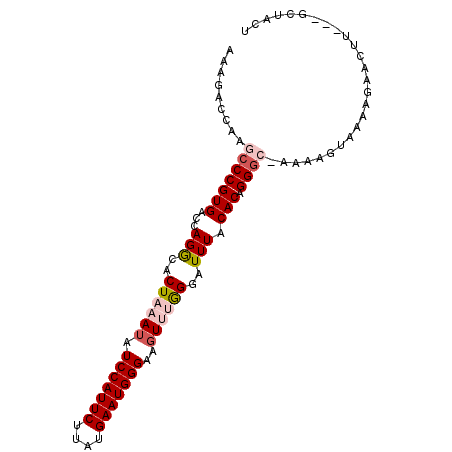
| Location | 8,857,144 – 8,857,237 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.02 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8857144 93 - 23771897 GAUUUACACAGGGCAAAAGUAAACGAACCUGCUACUCAAAUAUUGAGUUAUGUAGAUUUUCAGAA-----------------GAGAAAAUCAACUUCUACUGUGUAAAGG------ ..(((((((((.....((((.........(((.(((((.....)))))...)))(((((((....-----------------..))))))).))))...)))))))))..------ ( -24.60) >DroSec_CAF1 117047 110 - 1 GAUUUACACAGGGCAAAAGUAAAAGAACUUGCUACUUAAAUAUUGAGUUAUGUAGAUUUUCAAAAGAAGUUAGUUGGGUAAAGAGAAAAU------CUACUGUGUAAGCAGUUCUU (.(((((((((((((..(((......)))))))(((((.....)))))....(((((((((.......................))))))------)))))))))))))....... ( -21.90) >DroSim_CAF1 113475 116 - 1 GAUUUACACAGGGCAAAAGUAAAAGAACUUGCUACUUAAAUAUUGAGUUAUGUAGAUUUUCAAAAGAAGUUAGUUGGGUAAAGAGAAAAUCAACUUCUACUGUGUAAGCAGUACUU (.(((((((((((((..(((......))))))).........(((((...........))))).(((((((.(((............))).))))))).))))))))))....... ( -22.00) >consensus GAUUUACACAGGGCAAAAGUAAAAGAACUUGCUACUUAAAUAUUGAGUUAUGUAGAUUUUCAAAAGAAGUUAGUUGGGUAAAGAGAAAAUCAACUUCUACUGUGUAAGCAGU_CUU ..(((((((((.(((..(((((......)))))(((((.....)))))..))).(((((((.......................)))))))........)))))))))........ (-18.64 = -18.87 + 0.23)

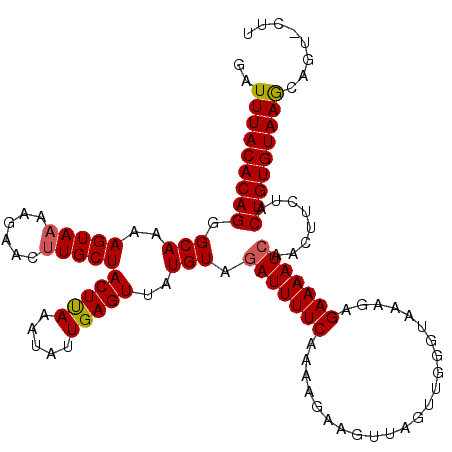

| Location | 8,857,201 – 8,857,294 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.95 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -16.16 |
| Energy contribution | -17.28 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8857201 93 - 23771897 AAAGACCAAGCCCGUGACCAGGCACUAAAUAUCCAUUCUUAUGAAUGGGAAGUUUAGGAUUUACACAGGGC-AAAAGUAAACGAACCU---GCUACU .........(((((((...(((..((((((.(((((((....)))))))..))))))..))).))).))))-...((((........)---)))... ( -24.00) >DroSec_CAF1 117121 93 - 1 AAAGACCAAGCCCGUGACCAGGCACUAAAUAUCCAUUCUUAUGAAUGGGAAGUCAGGGAUUUACACAGGGC-AAAAGUAAAAGAACUU---GCUACU .........(((((((...............(((((((....)))))))(((((...))))).))).))))-...(((((......))---)))... ( -22.40) >DroSim_CAF1 113555 93 - 1 AAAGACCAAGCCCGUGACCAGGCACUAAAUAUCCAUUCUUAUGAAUGGGAAGUCAGGGAUUUACACAGGGC-AAAAGUAAAAGAACUU---GCUACU .........(((((((...............(((((((....)))))))(((((...))))).))).))))-...(((((......))---)))... ( -22.40) >DroEre_CAF1 121367 92 - 1 GAAGACCAAGCCCGUGACCAGAGACUAAAAAUCCAUUCUUAUGAAUGGGAAGUUUAGCAUUUACACAGGC--AAAAGUAAAAGAACUU---GUAACU .........(((.(((...(((..(((((..(((((((....)))))))...)))))..))).))).)))--..((((......))))---...... ( -21.50) >DroYak_CAF1 120396 97 - 1 GAAGACCAAGCCCGUGAGCAGACACUAAAUAUCCAUUCUUAUGAAUGGGAAGUUUGGGAUUUACACAGGGAGAAAAGUCAACAAAAGUAGGGUAACG ...(((....((((((...(((..((((((.(((((((....)))))))..))))))..))).))).)))......))).................. ( -22.30) >consensus AAAGACCAAGCCCGUGACCAGGCACUAAAUAUCCAUUCUUAUGAAUGGGAAGUUUGGGAUUUACACAGGGC_AAAAGUAAAAGAACUU___GCUACU .........(((((((...(((..((((((.(((((((....)))))))..))))))..))).))).)))).......................... (-16.16 = -17.28 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:47 2006