
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,852,864 – 8,853,005 |
| Length | 141 |
| Max. P | 0.695154 |

| Location | 8,852,864 – 8,852,967 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -30.83 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
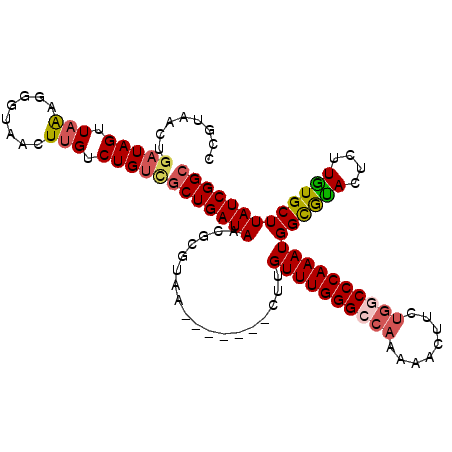
>3L_DroMel_CAF1 8852864 103 + 23771897 CCGUAACUGAUAGUUAAAGGGUAACUUGACUGUCGCUGAUAACGCGUUU-------CUUGUUUGGGCCAAAAACUUGUGUCCCAAAUGGUGUACUCUUGUGCUUAUCGGC ........(((((((((.(.....))))))))))((((((((.......-------.(..((((((((((....))).).))))))..).((((....)))))))))))) ( -31.00) >DroSec_CAF1 112899 102 + 1 CCGUAACUGAUAGUUAAAGGGUAACUUGUCUGUCGCUGAUAACGCGUUU-------CUUGUUUGGGCCAAAAAAUUCU-UCCCAAAUGGUGUACUCUUAUGCUUAUCGGC ........(((((...(((.....)))..)))))((((((((.((((..-------((..((((((............-.))))))..).).......)))))))))))) ( -25.42) >DroEre_CAF1 116697 110 + 1 CUGCAACUGAUAGUUAGCGGGUAAAUAGUCUGUGGCUGAUAACGGGUAACGGGUUCCUUGUUUGGGCCAAAAACUUCGGGCCCAAAUGGCGCACUCUUGUGCUUAUCGGC (((((((.....))).))))..............((((((((.(((........)))(..((((((((..........))))))))..).((((....)))))))))))) ( -35.50) >DroYak_CAF1 115920 97 + 1 ------CUGAUAGUUAAUGGCUAUCUUGUCUGCUACUGAUAACAAGGAA-------CUUGUUUGGGCCAAAAACUUCUGGCCCAAAUGGCAUACUCUUGUGCUUAUCGGC ------..((((((.....))))))..........((((((((((((..-------.(..(((((((((........)))))))))..).....))))))...)))))). ( -31.40) >consensus CCGUAACUGAUAGUUAAAGGGUAACUUGUCUGUCGCUGAUAACGCGUAA_______CUUGUUUGGGCCAAAAACUUCUGGCCCAAAUGGCGUACUCUUGUGCUUAUCGGC ........(((((.(((........))).)))))(((((((..................((((((((((........))))))))))((((((....))))))))))))) (-21.10 = -21.97 + 0.87)

| Location | 8,852,894 – 8,853,005 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -25.27 |
| Energy contribution | -26.40 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8852894 111 - 23771897 GGUGCAAGUAAAGGAAACGCGUGCCAUAGCAACGUCGCGCCGAUAAGCACAAGAGUACACCAUUUGGGACACAAGUUUUUGGCCCAAACAAG-------AAACGCGUUAUCAGCGACA (((((.......(....)((((((....)).)))).)))))(((((((((....))......((((((...(((....))).))))))....-------....)).)))))....... ( -28.40) >DroSec_CAF1 112929 110 - 1 GGUGCAAGUAAAGGAAACGCGUGCCAUCGCAACGUCGCGCCGAUAAGCAUAAGAGUACACCAUUUGGGA-AGAAUUUUUUGGCCCAAACAAG-------AAACGCGUUAUCAGCGACA (((((.......(....)((((((....)).)))).)))))(((((((....(.(....)).((((((.-.((.....))..))))))....-------....)).)))))....... ( -27.00) >DroEre_CAF1 116727 118 - 1 GGUGCAAGUAAAGGAAACGCGUGCCAUCGCAACGACGCGCCGAUAAGCACAAGAGUGCGCCAUUUGGGCCCGAAGUUUUUGGCCCAAACAAGGAACCCGUUACCCGUUAUCAGCCACA (((((.......(....)(((......)))......)))))(((((((((....)))).((.((((((((.((....)).))))))))...)).............)))))....... ( -37.70) >DroYak_CAF1 115944 111 - 1 GCUGCAAGUGGAGGAAACGCGUGGCAUCUCAACGACGCGCCGAUAAGCACAAGAGUAUGCCAUUUGGGCCAGAAGUUUUUGGCCCAAACAAG-------UUCCUUGUUAUCAGUAGCA (((((..(((((((((....((((((((((......((........))....))).)))))))((((((((((....)))))))))).....-------))))))..)))..))))). ( -42.00) >consensus GGUGCAAGUAAAGGAAACGCGUGCCAUCGCAACGACGCGCCGAUAAGCACAAGAGUACACCAUUUGGGACAGAAGUUUUUGGCCCAAACAAG_______AAACGCGUUAUCAGCGACA (((((.......(....)((((((....)).))).))))))(((((((......))......(((((((((((....)))))))))))..................)))))....... (-25.27 = -26.40 + 1.13)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:41 2006