
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,849,139 – 8,849,237 |
| Length | 98 |
| Max. P | 0.997016 |

| Location | 8,849,139 – 8,849,237 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -18.99 |
| Consensus MFE | -16.68 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
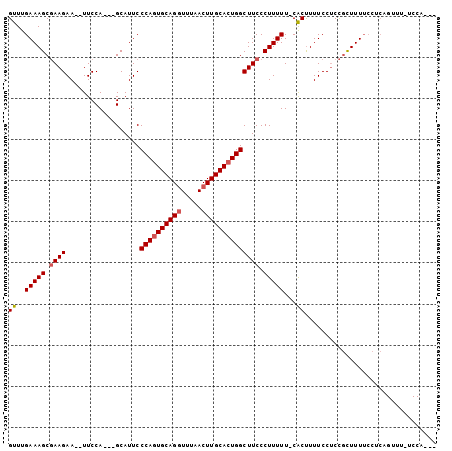
>3L_DroMel_CAF1 8849139 98 + 23771897 GUUUGAAAGCGAAGAA--UUCCA---GCAUUCCCAGUGCAGUUUUAACUUGCACUGGCUUCCCUUUUU-CACUUUUCCUCCGCUUUUCCUCAGUUUUUCCAGUU ....(((((((..(((--(....---..))))((((((((((....)).))))))))...........-...........)))))))................. ( -20.90) >DroSec_CAF1 109220 84 + 1 GUUUGAAAGCUAAGAA--UUCCAGCAGCAUUCCCAUUGCAGGUUUAACUUGCACUGGCUUCCCUUUUU-CACUUUUCCACUCCUUUU----------------- ...((((((((.....--....))).......(((.((((((.....)))))).)))........)))-))................----------------- ( -14.70) >DroEre_CAF1 113116 98 + 1 GUUUGAAAGCGAAGAACAUUCCA---GCAUUCCCAGUGCAGGUUCAACUUGCACUGGCUUCCCUUUUUCCGCCUUUCCUCCGUUUUGCCUCAGUUUCUCCA--- ((..(((((.((((.........---......((((((((((.....)))))))))))))).)))))...)).............................--- ( -21.36) >consensus GUUUGAAAGCGAAGAA__UUCCA___GCAUUCCCAGUGCAGGUUUAACUUGCACUGGCUUCCCUUUUU_CACUUUUCCUCCGCUUUUCCUCAGUUU_UCCA___ ((..(((((.((((..................((((((((((.....)))))))))))))).)))))...))................................ (-16.68 = -17.46 + 0.78)


| Location | 8,849,139 – 8,849,237 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -14.73 |
| Energy contribution | -15.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
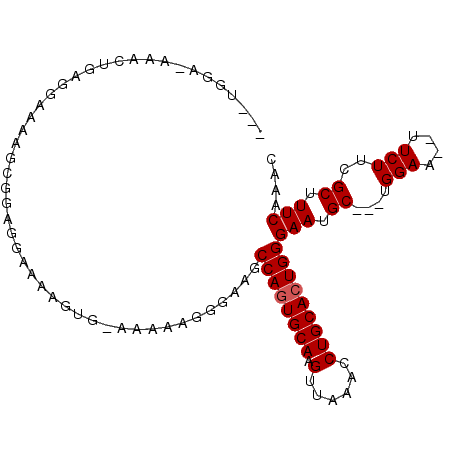
>3L_DroMel_CAF1 8849139 98 - 23771897 AACUGGAAAAACUGAGGAAAAGCGGAGGAAAAGUG-AAAAAGGGAAGCCAGUGCAAGUUAAAACUGCACUGGGAAUGC---UGGAA--UUCUUCGCUUUCAAAC ................((((.((((((((....(.-...).......((((((((.((....))))))))))......---.....--)))))))))))).... ( -25.50) >DroSec_CAF1 109220 84 - 1 -----------------AAAAGGAGUGGAAAAGUG-AAAAAGGGAAGCCAGUGCAAGUUAAACCUGCAAUGGGAAUGCUGCUGGAA--UUCUUAGCUUUCAAAC -----------------................((-(((.(((((..(((((...(((....((......))....))))))))..--)))))...)))))... ( -16.60) >DroEre_CAF1 113116 98 - 1 ---UGGAGAAACUGAGGCAAAACGGAGGAAAGGCGGAAAAAGGGAAGCCAGUGCAAGUUGAACCUGCACUGGGAAUGC---UGGAAUGUUCUUCGCUUUCAAAC ---.((((((.....((((...((.........))............((((((((.(.....).))))))))...)))---)......)))))).......... ( -24.40) >consensus ___UGGA_AAACUGAGGAAAAGCGGAGGAAAAGUG_AAAAAGGGAAGCCAGUGCAAGUUAAACCUGCACUGGGAAUGC___UGGAA__UUCUUCGCUUUCAAAC ...............................................((((((((.(......)))))))))(((.((....(((....)))..)).))).... (-14.73 = -15.07 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:39 2006