
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,847,957 – 8,848,118 |
| Length | 161 |
| Max. P | 0.995330 |

| Location | 8,847,957 – 8,848,059 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -18.03 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.995027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8847957 102 - 23771897 UUGCCACUU------CCCCUCCACAGCUAUUUUAACACUCCCCCCCACUCCUUCCGACGGGGCAUUUCCCGGUGGCAUAUGCAACUUGUAUGCGUUUUCAAUUUGCAC .((((((..------.................................((.....))(((((....)))))))))))..(((((.(((..........))).))))). ( -19.80) >DroSec_CAF1 108078 95 - 1 UUCCCACAU------CCCCCCCAUUGCUAUUUUG------CCCCCCUCUCC-CCCGCCGGGGCAUUUCCCGGUGGCAUAUGCAACUUGUAUGCGUUUUCAAUUUGCAC .........------.........(((....(((------...........-..((((((((....))))))))((((((.......)))))).....)))...))). ( -22.30) >DroYak_CAF1 111176 96 - 1 UCCCCAAUUACCACACUCCCCCAU------U-UAGCAAG-CCCCC---CUC-CCCGCAGAGACAUUUCCCAGUGGCAUAUGCAACUUGUAUGCGUUUUCAAUUUGCAC ........................------.-..(((((-.....---(((-......))).(((......)))((((((.......))))))........))))).. ( -12.00) >consensus UUCCCACUU______CCCCCCCAU_GCUAUUUUA_CA___CCCCCC_CUCC_CCCGCCGGGGCAUUUCCCGGUGGCAUAUGCAACUUGUAUGCGUUUUCAAUUUGCAC .....................................................(((((((((....)))))))))....(((((.(((..........))).))))). (-14.82 = -15.60 + 0.78)

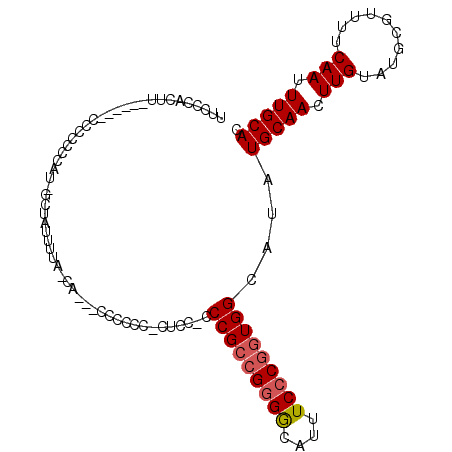

| Location | 8,848,025 – 8,848,118 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 72.64 |
| Mean single sequence MFE | -17.84 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.82 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8848025 93 - 23771897 UAUGGGAGUGGAUUUUGCAUACCUUGACAUAUUUCGAGUGUCCUUACUCCGCCACAUCCUUGCCACUU------CCCCUCCACAGCUAUUUUAACACUC ..((((((.(((....(((......((......))(((((....)))))...........)))....)------)).)))).))............... ( -17.30) >DroSec_CAF1 108145 87 - 1 UAUGGGAGUGGAUUUUGCAUACCUUGACAUAUUUCGAGUGUCCUUACUCCACCACAUCCUUCCCACAU------CCCCCCCAUUGCUAUUUUG------ ...((((((((....((((.....)).))......(((((....))))).............)))).)------)))................------ ( -16.40) >DroEre_CAF1 112124 77 - 1 CAUGGGAGUGGAUUGUGCAUACCUUGACAUAUUUCGAGUGUCCUUACUCCACCACAUCCAUCCC------CUCUCCCCACCAU---------------- ...(((..(((((.(((((.....)).........(((((....)))))...))))))))..))------)............---------------- ( -19.20) >DroYak_CAF1 111240 91 - 1 UAUGGGAGUGGAUUUUGCAUACCUUGACAUAUUUCGAGUGUCCUUACUCCACCACAGCCUCCCCAAUUACCACACUCCCCCAU------U-UAGCAAG- ...(((((((((...((((.....)).))....))(((((....))))).......................)))))))....------.-.......- ( -18.40) >DroAna_CAF1 108671 73 - 1 UUUGGGAGUGGAUUUUGCAUACCUUGACAUAUUUUGAGUGUCCUUACACCGCCACU----CCCCACUA------CUCUGGCUU---------------- ...((((((((....((((.....)).))......(.((((....))))).)))))----))).....------.........---------------- ( -17.90) >consensus UAUGGGAGUGGAUUUUGCAUACCUUGACAUAUUUCGAGUGUCCUUACUCCACCACAUCCUUCCCACUU______CCCCCCCAU______U_U_______ ..((((((((((.(..(.((((.((((......)))))))).)..).)))))........))))).................................. (-11.22 = -11.82 + 0.60)

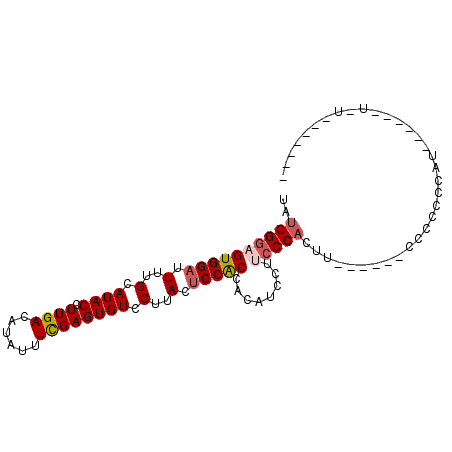
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:36 2006