
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,796,654 – 8,796,757 |
| Length | 103 |
| Max. P | 0.981682 |

| Location | 8,796,654 – 8,796,757 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.08 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8796654 103 - 23771897 CGUGCCAU---ACAGUAAGGCAUCCUGAAG----------UGAGGC--UCGCAACGUGUGCGAAUGUGUUGCACAAUAAAUAAAAAUAUUUAUACAUGAAACCUUAAAUAAAGCAUGA (((((...---....(((((..((.((..(----------..(.((--(((((.....)))))..)).)..)...(((((((....))))))).)).))..)))))......))))). ( -24.82) >DroVir_CAF1 68209 106 - 1 CGUGCUGC---AC--------AUCCUGCAGCUCGCUGGC-AGACGUAGCUGCAACGUGUGCGAAUGUGUUGCACAAUAAAUAAAAAUAUUUAUACAUGAAACCCUAAAUAAAGCAUGA ((((((((---((--------((..((((((((((....-.).)).)))))))..))))))...((((...))))(((((((....)))))))..................)))))). ( -32.30) >DroGri_CAF1 59809 102 - 1 CGUGCUGA---AC--------AUCCUGCAGCUCGAC-----GAUGCUGCUGCAACGUGUGCGAAUGUGUUGCACAAUAAAUAAAAAUAUUUAUACAUGAAACCCUAAAUAAAGCAUGA ((((((..---..--------.....(((((.....-----...)))))(((((((..(....)..)))))))..(((((((....)))))))..................)))))). ( -27.50) >DroWil_CAF1 74819 108 - 1 CGUGCCGU---ACA------CAUCCUGAAGCACUUUGGAGUAGACCU-GCACAACGUGUGCGAAUGUGUUGCACAAUAAAUAAAAAUAUUUAUACAUGAAACCUUAAAUAAAGCAUGA .((((...---(((------(((.(....((((.(((..(((....)-)).))).))))..).)))))).)))).(((((((....))))))).((((...............)))). ( -23.06) >DroAna_CAF1 59660 103 - 1 CGUGCCUC---ACCGUAUGGCAUCCCGAGC----------CUCAGC--UCGCAACGUGUGCGAAUGUGUUGCACAAUAAAUAAAAAUAUUUAUACAUGAAACCUUAAAUAAAGCAUGA .(((((..---.......)))))...((((----------....))--))((((((..(....)..))))))...(((((((....))))))).((((...............)))). ( -23.96) >DroPer_CAF1 67069 97 - 1 CGUGCAUCCUCUCUCUCUCUCUUUCUG---------------------UGGCAACGUGUGCGAAUGUGUUGCACAAUAAAUAAAAAUAUUUAUACAUGAAACCUUAAAUAAAGCAUGA (((((....................((---------------------((.(((((..(....)..)))))))))(((((((....)))))))...................))))). ( -19.40) >consensus CGUGCCGC___AC_______CAUCCUGAAG___________GAAGC__UCGCAACGUGUGCGAAUGUGUUGCACAAUAAAUAAAAAUAUUUAUACAUGAAACCUUAAAUAAAGCAUGA (((((.............................................((((((..(....)..))))))...(((((((....)))))))...................))))). (-16.48 = -16.65 + 0.17)

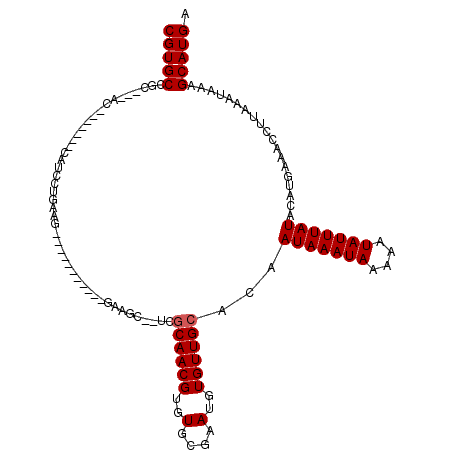
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:24 2006