| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
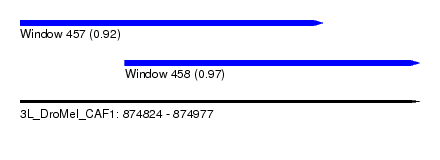
| Location | 874,824 – 874,977 |
| Length | 153 |
| Max. P | 0.970904 |

| Location | 874,824 – 874,940 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.98 |
| Mean single sequence MFE | -47.63 |
| Consensus MFE | -38.88 |
| Energy contribution | -36.50 |
| Covariance contribution | -2.38 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
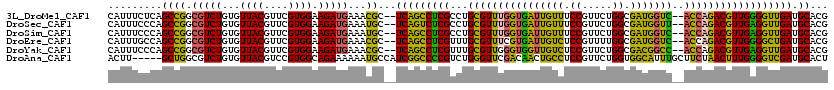
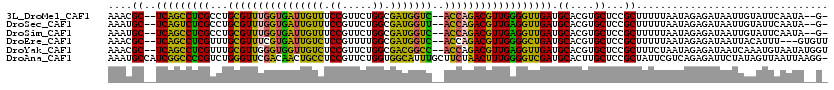
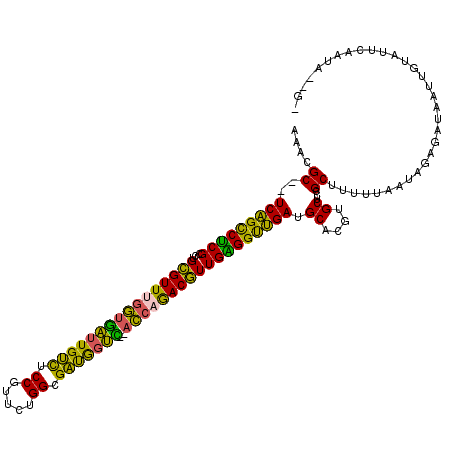
>3L_DroMel_CAF1 874824 116 + 23771897 CAUUUCUCAGCCGGCGUCUGUGUUACGUUCGUGGAAGAUGAAACGC--UCAGCCUCGCCUGCGUUUGGUGAUUGUUUCCGUUCUGGCGAUGGUC--ACCAGACGUUGGGGUUGAUGCACG (((((..((.(..((((.......))))..)))..)))))....((--(((((((((...((((((((((((..((.((.....)).))..)))--)))))))))))))))))).))... ( -48.00) >DroSec_CAF1 17717 116 + 1 CAUUUCCCAGCCGGCGUCUGUGUUACGUUCGUGGAAGAUGAAAUGC--UCAGUCUCGCCUGCGUUUGGUGAUUGUUUCCGUUCUGGCGAUGGUU--ACCAGACGUUGAGGUUGAUGCACG (((((.(((.(..((((.......))))..)))).)))))...(((--(((..((((...((((((((((((..((.((.....)).))..)))--)))))))))))))..))).))).. ( -43.90) >DroSim_CAF1 12485 116 + 1 CAUUUCCCAGCCGGCGUCUGUGUUACGUUCGUGGAAGAUGAAAUGC--UCAGCCUCGCCUGCGUUUGGUGAUUGUUUCCGUUCUGGCGAUGGUC--ACCAGACGUUGAGGUUGAUGCACG (((((.(((.(..((((.......))))..)))).)))))...(((--(((((((((...((((((((((((..((.((.....)).))..)))--)))))))))))))))))).))).. ( -50.40) >DroEre_CAF1 17795 116 + 1 CAUUUGCCAGCCGGCGUCUGUGUUACGUUCGUGGAAGAUGAAACGC--UCAGCCUCGUUUGCGUUUCGUGAUUGUCUCCGUUUUGGCGAUGGUC--ACCAGACGUUGGGGCUGAUGCACG (((((.(((.(..((((.......))))..)))).)))))....((--(((((((((...((((((.(((((..((.((.....)).))..)))--)).))))))))))))))).))... ( -47.20) >DroYak_CAF1 17843 116 + 1 CAUUUCCCAGCCGGCGUCUGUGUUACGUUCGUGGAAGAUGAAACGC--UCAGCCUCGUUUGCGUUGGGUGGUUGUCUCCGUUCUGGCGACGGCC--ACCAGACGUUGAGGUUGAUGCACG (((((.(((.(..((((.......))))..)))).)))))....((--(((((((((...(((((.((((((((((.((.....)).)))))))--))).)))))))))))))).))... ( -52.00) >DroAna_CAF1 16950 115 + 1 ACUU-----GCUGGCGUCUGUGUUACGUCCGUGGCAGAAAAAAUGCCAUCGGCCCCGUCUGGGUUCGACAACUGCCUCCGUUCUGGUGGCAUUUGCUUCUAACUUUGGGGUCGAUGCACU ...(-----((.(((((.(.((((((....)))))).)....)))))((((((((((...(((((.(((((.((((.((.....)).)))).)))..)).)))))))))))))))))).. ( -44.30) >consensus CAUUUCCCAGCCGGCGUCUGUGUUACGUUCGUGGAAGAUGAAACGC__UCAGCCUCGCCUGCGUUUGGUGAUUGUCUCCGUUCUGGCGAUGGUC__ACCAGACGUUGAGGUUGAUGCACG .........((((.(((((...((((....)))).)))))...))...(((((((((...((((((((((((((((.((.....)).)))))))..)))))))))))))))))).))... (-38.88 = -36.50 + -2.38)
| Location | 874,864 – 874,977 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.23 |
| Mean single sequence MFE | -40.71 |
| Consensus MFE | -32.97 |
| Energy contribution | -30.65 |
| Covariance contribution | -2.32 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 874864 113 + 23771897 AAACGC--UCAGCCUCGCCUGCGUUUGGUGAUUGUUUCCGUUCUGGCGAUGGUC--ACCAGACGUUGGGGUUGAUGCACGUGCUCCGCUUUUUAAUAGAGAUAAUUGUAUUCAAUA--G- ....((--(((((((((...((((((((((((..((.((.....)).))..)))--)))))))))))))))))).))..((((....((((.....))))......))))......--.- ( -41.70) >DroSec_CAF1 17757 113 + 1 AAAUGC--UCAGUCUCGCCUGCGUUUGGUGAUUGUUUCCGUUCUGGCGAUGGUU--ACCAGACGUUGAGGUUGAUGCACGUGCUCCGCUUUUUAAUAGAGAUAAUUGUAUUCAAUA--G- ...(((--(((..((((...((((((((((((..((.((.....)).))..)))--)))))))))))))..))).))).((((....((((.....))))......))))......--.- ( -36.50) >DroSim_CAF1 12525 113 + 1 AAAUGC--UCAGCCUCGCCUGCGUUUGGUGAUUGUUUCCGUUCUGGCGAUGGUC--ACCAGACGUUGAGGUUGAUGCACGUGCUCCGCUUUUUAAUAGAGAUAAUUGUAUUCAAUA--G- ...(((--(((((((((...((((((((((((..((.((.....)).))..)))--)))))))))))))))))).))).((((....((((.....))))......))))......--.- ( -43.00) >DroEre_CAF1 17835 113 + 1 AAACGC--UCAGCCUCGUUUGCGUUUCGUGAUUGUCUCCGUUUUGGCGAUGGUC--ACCAGACGUUGGGGCUGAUGCACGUGCUCCGCUUUUUAAUAGAGAUAAUUACAUUU---GUGUU ....((--(((((((((...((((((.(((((..((.((.....)).))..)))--)).))))))))))))))).))..(((.....((((.....)))).....)))....---..... ( -39.80) >DroYak_CAF1 17883 116 + 1 AAACGC--UCAGCCUCGUUUGCGUUGGGUGGUUGUCUCCGUUCUGGCGACGGCC--ACCAGACGUUGAGGUUGAUGCACGUGCUCCGCUUUCUAAUAGAGAUAAUCAAAUGUAAUAUGGU ....((--(((((((((...(((((.((((((((((.((.....)).)))))))--))).)))))))))))))).))((((.......((((.....)))).......))))........ ( -45.04) >DroAna_CAF1 16985 119 + 1 AAAUGCCAUCGGCCCCGUCUGGGUUCGACAACUGCCUCCGUUCUGGUGGCAUUUGCUUCUAACUUUGGGGUCGAUGCACUUGCUCCGCUAUUCGUCAGAGAUUCUAUAGUUAAUUAAGG- ...(((.((((((((((...(((((.(((((.((((.((.....)).)))).)))..)).))))))))))))))))))((((....(((((..(((...)))...)))))....)))).- ( -38.20) >consensus AAACGC__UCAGCCUCGCCUGCGUUUGGUGAUUGUCUCCGUUCUGGCGAUGGUC__ACCAGACGUUGAGGUUGAUGCACGUGCUCCGCUUUUUAAUAGAGAUAAUUGUAUUCAAUA__G_ ....((..(((((((((...((((((((((((((((.((.....)).)))))))..)))))))))))))))))).((....))...))................................ (-32.97 = -30.65 + -2.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:49 2006