
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,760,521 – 8,760,629 |
| Length | 108 |
| Max. P | 0.500000 |

| Location | 8,760,521 – 8,760,629 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.33 |
| Mean single sequence MFE | -16.28 |
| Consensus MFE | -10.42 |
| Energy contribution | -10.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
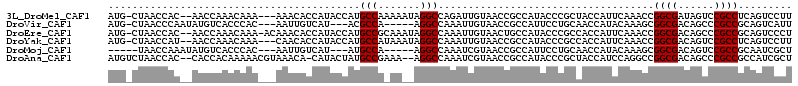
>3L_DroMel_CAF1 8760521 108 + 23771897 AUG-CUAACCAC--AACCAAACAAA---AAACACCAUACCAUGCCAAAAAUAGGCCAGAUUGUAACCGCCAUACCCGCUACCAUUCAAACCGGCGAUAGUCCGCCUCAGUCCUU ...-........--...........---..............(((.......)))..(((((.....((.......)).............((((......)))).)))))... ( -13.30) >DroVir_CAF1 33897 102 + 1 AUG-CUAACCCAAUAUGUCACCCAC---AAUUGUCAU---ACGCCA-----AGGCCAAAUUGUAACCGCCAUUCCUGCAACCAUACAAAGCGGCGACAGCCCGCCGCAGUCAUU .((-(........((((.((.....---...)).)))---).....-----.(((............)))......)))..........((((((......))))))....... ( -20.60) >DroEre_CAF1 27628 110 + 1 AUG-CUAACCAC--AACCAAACAAA-ACAAACACCAUACCAUGCCGCAAAUAGGCCAAAUUGUAACUGCCAUACCCGCCACCAUUCAAACCGGCGACAGCCCGCCGCAGUCCCU ...-........--...........-................(((.......)))......(..(((((.......(....).........((((......)))))))))..). ( -15.20) >DroYak_CAF1 27792 108 + 1 AUG-CUAACCAU--AACCAAACAAA---CAACACCAUACCAUGCCAUAAAUAGGCCAAAUUGUAACCGCCAUACCCGCCACCAUUCAAACCGGCGACAGUCCGCCUCAGUCCUU ...-........--...........---.......................((((...(((((...((((.....................)))))))))..))))........ ( -11.50) >DroMoj_CAF1 32095 98 + 1 -----UAACCAAAUAUGUCACCCAC---AAUUGUCAU---AUGCCA-----AGGCCAAAUCGUAACCGCCAUUCCUGCAACCAUACAAAGCGGCGACAGUCCGCCGCAAUCGCU -----.......(((((.((.....---...)).)))---))((..-----.(((............)))...................((((((......))))))....)). ( -20.30) >DroAna_CAF1 26484 109 + 1 AUGUCUAACCAC--CACCACAAAAACGUAAACA-CAUACUAUGCCGAAA--AGGCCAAAUCGUAACCGCCAUACCCGCUACCAUCCAGGCCGGCGACAGCCCGCCGCCAUCGCU ............--............(((....-........(((....--.)))............((.......)))))......(((.((((......)))))))...... ( -16.80) >consensus AUG_CUAACCAC__AACCAAACAAA___AAACACCAUACCAUGCCA_AA__AGGCCAAAUUGUAACCGCCAUACCCGCAACCAUUCAAACCGGCGACAGCCCGCCGCAGUCCCU ..........................................(((.......)))....................................((((......))))......... (-10.42 = -10.42 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:17 2006