| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,748,584 – 8,748,690 |
| Length | 106 |
| Max. P | 0.935799 |

| Location | 8,748,584 – 8,748,690 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.01 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -20.84 |
| Energy contribution | -22.90 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
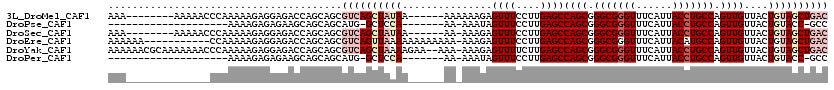
>3L_DroMel_CAF1 8748584 106 + 23771897 AAA--------AAAAACCCAAAAAGAGGAGACCAGCAGCGUCAGCUAUAA------AAAAAAGAGUUUCCUUGAGCCAGCGGGCGGGUUUCAUUACCUGCCAGUUGUUACUGUAGCUGAC ...--------.............(.(....))......((((((((((.------........((((....))))((((.(((((((......))))))).))))....)))))))))) ( -31.90) >DroPse_CAF1 16624 90 + 1 --------------------AAAAGAGAGAAGCAGCAGCAUG-GCUCCA-------AA-AAAUAGUUUCCUUGAGCCAGCGGGCGGGUUUCAUUACCUGCCAGUUGUUACUGUACC-GCC --------------------.....(.((.((((((.((.((-((((..-------..-.............)))))))).(((((((......))))))).)))))).)).)...-... ( -31.03) >DroSec_CAF1 15137 105 + 1 AAA--------AAAAACCCAAAAAGAGGAGACCAGCAGCGUCAGCUAUAA------AA-AAAGAGUUUCCUUGAGCCAGCGGGCGGGUUUCAUUACCUGCCAGUUGUUACUGUAGCUGAC ...--------.............(.(....))......((((((((((.------..-.....((((....))))((((.(((((((......))))))).))))....)))))))))) ( -31.90) >DroEre_CAF1 15442 108 + 1 AAAAAA-----------CCAAAAAGAGGAGACCAGCAGCGUCAGUUAAAAAAAAAAAA-AAAGAGUUUCCUUGAGCCAGCGGGCGGGUUUCAUUACAUGCCAGUUGUUACUGUAGCUGAC ......-----------.......(.(....))..((((..((((.............-.....((((....))))((((.((((.((......)).)))).))))..))))..)))).. ( -20.90) >DroYak_CAF1 15460 117 + 1 AAAAAACGCAAAAAAACCCAAAAAGAGGAGACCAGCAGCGUCAGCUAAAAGAA--AAA-AAAGAGUUUUCUUGAGCCAGCGGGCGGGUUUCAUUACCUGCCAGUUGUUACUGUAGCUGAC ........................(.(....))......((((((((.(((((--((.-......)))))))....((((.(((((((......))))))).))))......)))))))) ( -32.80) >DroPer_CAF1 16174 90 + 1 --------------------AAAAGAGAGAAGCAGCAGCAUG-GCUCCA-------AA-AAAUAGUUUCCUUGAGCCAGCGGGCGGGUUUCAUUACCUGCCAGUUGUUACUGUACC-GCC --------------------.....(.((.((((((.((.((-((((..-------..-.............)))))))).(((((((......))))))).)))))).)).)...-... ( -31.03) >consensus AAA______________CCAAAAAGAGGAGACCAGCAGCGUCAGCUAAAA______AA_AAAGAGUUUCCUUGAGCCAGCGGGCGGGUUUCAUUACCUGCCAGUUGUUACUGUAGCUGAC .......................................((((((((((...............((((....))))((((.(((((((......))))))).))))....)))))))))) (-20.84 = -22.90 + 2.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:14 2006