
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,727,498 – 8,727,596 |
| Length | 98 |
| Max. P | 0.517838 |

| Location | 8,727,498 – 8,727,596 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.57 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
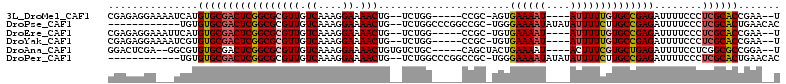
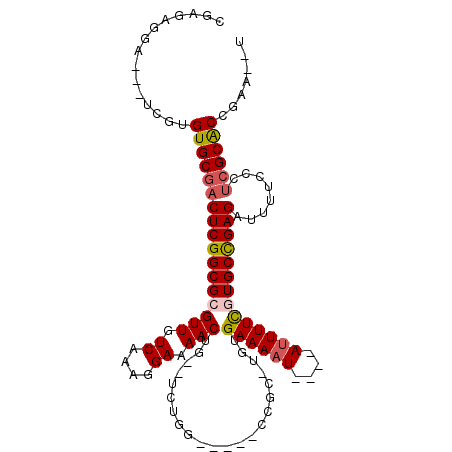
>3L_DroMel_CAF1 8727498 98 - 23771897 CGAGAGGAAAAUCAUGUGCGACUCGGCGCGUUGUCAAAGGAAAACUG--UCUGG-----CCGC-AGUGAAAAU----AUUUUUGUGCCGAGAUUUUCCCUCGCACCGAA--U ((((.((((((((..(.....)(((((((((.((((.((.....)).--..)))-----).))-.((....))----......))))))))))))))))))).......--. ( -31.00) >DroPse_CAF1 11573 97 - 1 ------------UGUGUGCGACUCGGCGCGUUGUCAAAGGAAAACUG--UCUGGCCCGGCCGC-UGGGAAAAUAUAUAUUUUCUUGCCGAGAUUUUCCCUCGCACUGAACAC ------------...(((((((((((((((((.((....)).)))..--...(((...)))))-.((((((((....))))))))))))))........))))))....... ( -29.40) >DroEre_CAF1 10280 98 - 1 CGAGAGGAAAUUCAUGUGCGACUCGGCGCGUUGUCAAAGGAAAACUG--UCUGG-----CCGC-UGUGAAAAU----AUUUUUGUGCCGAGAUUUUCCCUCGCACCGAA--U ((((.(((((.((......))((((((((((.((((.((.....)).--..)))-----).))-(((....))----).....))))))))..))))))))).......--. ( -29.70) >DroYak_CAF1 8506 98 - 1 CGAGAGGAAAAUCGUGUGCGACUCGGCGCGUUGUCAAAGGAAAACUG--UCUGG-----CCGC-UGUGAAAAU----AUUUUUGUGCCGAGAUUUUCCCUCGCACCGAA--U ((((.(((((((((....)))((((((((((.((((.((.....)).--..)))-----).))-(((....))----).....)))))))).)))))))))).......--. ( -33.70) >DroAna_CAF1 8445 99 - 1 GGACUCGA--GGCGUGUGCGACUCGGCGCGUUGUCAAAGGAAAACUGUGUCUGC-----CAGCUACUGAAAAU----ACUUUCGUGCUGAGAUUUUCCUCGGCGCCGGA--U ((..((((--((((....)).((((((((((((.((.((.....)).....)).-----))))....((((..----..)))))))))))).....))))))..))...--. ( -29.60) >DroPer_CAF1 11620 97 - 1 ------------UGUGUGCGACUCGGCGCGUUGUCAAAGGAAAACUG--UCUGGCCCGGCCGC-UGGGAAAAUAUAUAUUUUCUUGCCGAGAUUUUCCCUCGCACUGAACAC ------------...(((((((((((((((((.((....)).)))..--...(((...)))))-.((((((((....))))))))))))))........))))))....... ( -29.40) >consensus CGAGAGGA___UCGUGUGCGACUCGGCGCGUUGUCAAAGGAAAACUG__UCUGG_____CCGC_UGUGAAAAU____AUUUUCGUGCCGAGAUUUUCCCUCGCACCGAA__U ...............(((((((((((((((((.((....)).)))......................((((((....))))))))))))))........))))))....... (-21.64 = -21.95 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:10 2006