| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 870,502 – 870,613 |
| Length | 111 |
| Max. P | 0.539110 |

| Location | 870,502 – 870,613 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
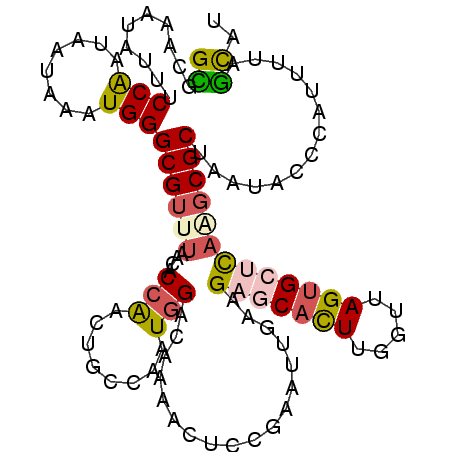
>3L_DroMel_CAF1 870502 111 + 23771897 GCGCAAAUAUUUCCAAUAAUAAAUGGGCGUUUAACACCAACUGCCAAUGGACAAAACUCCAAAUUGAAGAGCACUUGGUUAGUGCUCAAGCGCUAAUACACAUUUUAGCAU ..((.........((((........((((((.......))).)))..((((......)))).))))..(((((((.....)))))))..))(((((........))))).. ( -28.20) >DroSec_CAF1 13408 111 + 1 GCGGAAAUAUUUCCAAUAAUAAAUGGGCGUUUAACACCAACUGGCAAUGGACAAAACUCCGAAUAGAAGAGCACUUGGUUAGUGCUCAAGCGCUAAUAUCCAUUUUAGCAU ((((((....))))......((((((((((((........(((....((((......))))..)))..(((((((.....)))))))))))))......))))))..)).. ( -29.80) >DroSim_CAF1 8170 111 + 1 GCGAAAAUAUUUCCAAUAAUAAAUGGGCGUUUAACACCAACUGGCAAUGGACAAAACUCCGAAUUGAAGAGCACUUGGUUAGUGGUCAAGCGCUAAUAUCCAUUUUAGCAU ((.(((((....(((........)))((((((...((((.(((((..((((......)))).....(((....))).))))))))).))))))........))))).)).. ( -22.90) >DroEre_CAF1 13393 111 + 1 GUGCACGUAAUUCCAAUCAUAAAUGGGCGUUUAACACCGGCUGCCAAUGGCCAAAACUUCGAAUUGAAGUGCACUUGGUUAGAGCUCAGGCGCUAAUUCCCAUUUUAGCAU ((((((.((((((............((.(.....).))((((......))))........))))))..)))))).........((....))(((((........))))).. ( -28.00) >DroYak_CAF1 13452 111 + 1 GUGGAAAUAAUUCCAAUAAUAAAUGGGCGCCUAACACCGGCUGCCAAUGGCCAAAACUCUGAAUUGAAGAGCACUUGGUUAGUGCUCAAUCGCUAAUUCCCAUUUUAGCAA .(((((....)))))........(((((((.((((...((((......))))....((((.......))))......)))))))))))...(((((........))))).. ( -31.60) >DroAna_CAF1 12921 95 + 1 GUG--------UCCG---AUAAAUGGGCGUUUAGCACCGCCUCCCAAUUGUCGGU---CC-A-GCCGAGGGCAUUCAGUUAGUGGAUCUGCGCUAAUUCUAAUUUAAAUAG (((--------(((.---......(((((........)))))........((((.---..-.-.))))))))))..((((((((......))))))))............. ( -25.40) >consensus GCGCAAAUAUUUCCAAUAAUAAAUGGGCGUUUAACACCAACUGCCAAUGGACAAAACUCCGAAUUGAAGAGCACUUGGUUAGUGCUCAAGCGCUAAUACCCAUUUUAGCAU ((..........(((........)))((((((....(((........)))..................(((((((.....)))))))))))))..............)).. (-14.77 = -15.17 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:47 2006