
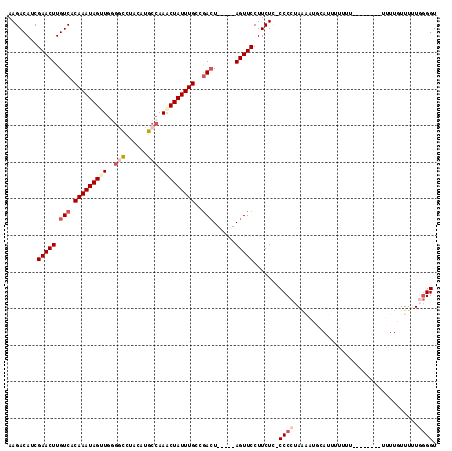
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,709,104 – 8,709,207 |
| Length | 103 |
| Max. P | 0.822169 |

| Location | 8,709,104 – 8,709,207 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -14.52 |
| Energy contribution | -15.96 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8709104 103 + 23771897 AAGACAUCGAACUUGUCACAAAUAGUUGGGACAUACAUGCCAAGCUAUUUGCCGACU-----AGUUCCUUCUC-CCCCUAAAAUGCAUUUUUGU--------UUUUGUUUUUGGGGU ........(((((.(((.(((((((((.((.((....)))).)))))))))..))).-----)))))......-.((((((((.(((.......--------...))))))))))). ( -29.90) >DroSec_CAF1 14603 100 + 1 AAGACAUCGAACUUGUCACAAAUAGUUGGGGCUUACAUGCCAAACUAUUUGCCGACU-----AGUUCCUUCUC-CCCCUA---UGCAUUUGUUU--------UUUUGUUUUUUGGGU ........(((((.(((.(((((((((..(((......))).)))))))))..))).-----)))))......-.(((..---.(((.......--------...))).....))). ( -28.20) >DroSim_CAF1 15429 102 + 1 AAGACAUCGAACUUGUCACAAAUAGUUGGGGCCUACAUGCCAAACUAUUUGCCGACU-----AGUUCCUUCUC-CCCCUAAAAUGCAUUUGUUU---------UUUGUUUUUGGGGU ........(((((.(((.(((((((((..(((......))).)))))))))..))).-----)))))......-.((((((((.(((.......---------..))))))))))). ( -32.30) >DroEre_CAF1 14449 101 + 1 AAGACAUCGAACUUGUCACAAAUAGCUGUUGCCUACAUGCCAAACUAUUUGCCGACU-----AGUUCCUUCUC-CCCCUAAAUUUAAUUUUU-U--------GGUU-UUUGUGGGGU ........(((((.(((.(((((((...(((.(.....).))).)))))))..))).-----))))).....(-(((.((((.((((....)-)--------))..-)))).)))). ( -23.10) >DroYak_CAF1 14699 107 + 1 AAGACAUCGAACUUGUCACAAAUAGCUGCGUCUUACAUGCCAAACUAUUUGCCGACUAUGUUAGUUCCUUCUG-CCCCUAAAUUUAAUUUUU-U--------UGUUGUUUCUGGGGU .(((....(((((.(((.(((((((..((((.....))))....)))))))..)))......)))))..)))(-((((......((((....-.--------.)))).....))))) ( -22.30) >DroAna_CAF1 14111 112 + 1 AAGACAUCGAACUUGUCACAAAUAGUUUGGGUCUGCAUGCCAAACUAUUUGCUAAGU-----AGUUCCUUCUAGCCCCUAGUAUUUAUCUAUUUUUUCUCUAUUUUUUUUUUUUGGU ..((((.......)))).((((((((((((.........))))))))))))..((((-----((......((((...)))).......))))))....................... ( -21.52) >consensus AAGACAUCGAACUUGUCACAAAUAGUUGGGGCCUACAUGCCAAACUAUUUGCCGACU_____AGUUCCUUCUC_CCCCUAAAAUGCAUUUUUUU________UUUUGUUUUUGGGGU ........(((((.(((.(((((((((..(((......))).)))))))))..)))......))))).......((((..................................)))). (-14.52 = -15.96 + 1.44)


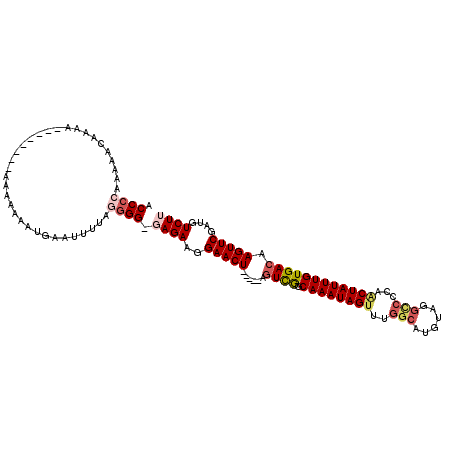
| Location | 8,709,104 – 8,709,207 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.65 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -17.03 |
| Energy contribution | -18.15 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8709104 103 - 23771897 ACCCCAAAAACAAAA--------ACAAAAAUGCAUUUUAGGGG-GAGAAGGAACU-----AGUCGGCAAAUAGCUUGGCAUGUAUGUCCCAACUAUUUGUGACAAGUUCGAUGUCUU .((((.((((.....--------...........)))).))))-((((..(((((-----.((((.(((((((.((((.((....)).))))))))))))))).)))))....)))) ( -29.19) >DroSec_CAF1 14603 100 - 1 ACCCAAAAAACAAAA--------AAACAAAUGCA---UAGGGG-GAGAAGGAACU-----AGUCGGCAAAUAGUUUGGCAUGUAAGCCCCAACUAUUUGUGACAAGUUCGAUGUCUU .(((...........--------...........---..)))(-((.(..(((((-----.((((.(((((((((.(((......)))..))))))))))))).)))))..).))). ( -28.06) >DroSim_CAF1 15429 102 - 1 ACCCCAAAAACAAA---------AAACAAAUGCAUUUUAGGGG-GAGAAGGAACU-----AGUCGGCAAAUAGUUUGGCAUGUAGGCCCCAACUAUUUGUGACAAGUUCGAUGUCUU .((((.((((....---------...........)))).))))-((((..(((((-----.((((.(((((((((.(((......)))..))))))))))))).)))))....)))) ( -31.96) >DroEre_CAF1 14449 101 - 1 ACCCCACAAA-AACC--------A-AAAAAUUAAAUUUAGGGG-GAGAAGGAACU-----AGUCGGCAAAUAGUUUGGCAUGUAGGCAACAGCUAUUUGUGACAAGUUCGAUGUCUU .((((.....-....--------.-..............))))-((((..(((((-----.((((.(((((((((..((......))...))))))))))))).)))))....)))) ( -29.06) >DroYak_CAF1 14699 107 - 1 ACCCCAGAAACAACA--------A-AAAAAUUAAAUUUAGGGG-CAGAAGGAACUAACAUAGUCGGCAAAUAGUUUGGCAUGUAAGACGCAGCUAUUUGUGACAAGUUCGAUGUCUU .((((.(((......--------.-..........))).))))-.(((..(((((......((((.((((((((...((.........)).)))))))))))).)))))....))). ( -20.93) >DroAna_CAF1 14111 112 - 1 ACCAAAAAAAAAAAAUAGAGAAAAAAUAGAUAAAUACUAGGGGCUAGAAGGAACU-----ACUUAGCAAAUAGUUUGGCAUGCAGACCCAAACUAUUUGUGACAAGUUCGAUGUCUU ...........................(((((....((((...))))...(((((-----.....(((((((((((((.........)))))))))))))....)))))..))))). ( -24.80) >consensus ACCCCAAAAACAAAA________AAAAAAAUGAAUUUUAGGGG_GAGAAGGAACU_____AGUCGGCAAAUAGUUUGGCAUGUAGGCCCCAACUAUUUGUGACAAGUUCGAUGUCUU .((((..................................)))).((((..(((((......((((.((((((((..(((......)))...)))))))))))).)))))....)))) (-17.03 = -18.15 + 1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:05 2006