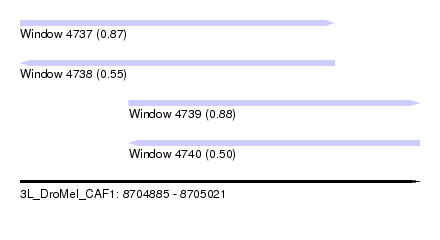
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 8,704,885 – 8,705,021 |
| Length | 136 |
| Max. P | 0.878433 |

| Location | 8,704,885 – 8,704,992 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -40.35 |
| Consensus MFE | -29.82 |
| Energy contribution | -30.78 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8704885 107 + 23771897 UUC-UGUAUUCUGGCUGGUAAGCAAUCGCAUC--UGCUGACUUGUUUGGGGCCA---AACUCUUGGCCAAGAGCUUCAAUGUUGCUGGCCAUC---GCUUGACAUUCG--AGUCGAGC-- ...-.......((((..(((((((........--)))(((...((((..(((((---(....))))))..)))).)))...))))..))))..---(((((((.....--.)))))))-- ( -39.00) >DroSec_CAF1 10432 108 + 1 UUUAUAUUUUCUGGCUGGUAAGCAAUCGCAUC--UGCUGACUUGUUUGGGGCCA---AACUCUUGGCCACGAGCUUCAAUGCUGCUGGCCAUC---GCUUGACAUUCG--AGUCGAGC-- ...........((((..(((.(((...((...--.))(((...(((((.(((((---(....)))))).))))).))).))))))..))))..---(((((((.....--.)))))))-- ( -42.30) >DroSim_CAF1 11249 108 + 1 UUCAUAUUUUCUGGCUGGUAAGCAAUCGCAUC--UGCUGACUUGUUUGGGGCCA---AACUCUUGGCCAAGAGCUUCAAUGCUGCUGGCCAUC---GCUUGACAUUCG--AGUCGAGC-- ...........((((..(((.(((...((...--.))(((...((((..(((((---(....))))))..)))).))).))))))..))))..---(((((((.....--.)))))))-- ( -41.30) >DroEre_CAF1 10415 105 + 1 ---AUAUUUUCUGGCUGCUAAGCAAUCGCAUC--UGCUGACUUGUUUGGGGCCG---AACUCUUGGCCAAGAGCUUCAAUGCUGCUGGCCAGC---GCUUGACAUUCG--AGUCGAGC-- ---.....(((.((((.((((((((((((...--.)).)).)))))))))))))---))...(((((((..(((......)))..))))))).---(((((((.....--.)))))))-- ( -40.80) >DroYak_CAF1 10533 107 + 1 ---AUAUUUUCUGGCUGCUAAGCAAUCGCAUC--UGCUGACUUGUUUGGGGCCA---AACUCUUGGCCAAGAGCUUCAAUGCUGCUGGCCAUC---GCUUGACAUUCGACAGUCGAGC-- ---........(((((.((((((((((((...--.)).)).)))))))))))))---......((((((..(((......)))..))))))..---(((((((........)))))))-- ( -40.60) >DroAna_CAF1 10262 107 + 1 UUCUGAUUUUCUGGCUGGUAAGCAAACCGUUCUGUGCUGAUUUCUGUUUGGCCAGAAAACUCUUGGCCAU-----------UUCUUGGCCAUCCAAGCUUGACAUUCG--AGUCGAGCUG ....(((((((((((..(..((((..........)))).........)..))))))))).)).((((((.-----------....))))))....((((((((.....--.)))))))). ( -38.10) >consensus UUCAUAUUUUCUGGCUGGUAAGCAAUCGCAUC__UGCUGACUUGUUUGGGGCCA___AACUCUUGGCCAAGAGCUUCAAUGCUGCUGGCCAUC___GCUUGACAUUCG__AGUCGAGC__ ...........(((((...((((((((((......)).)).))))))..))))).........((((((..(((......)))..)))))).....(((((((........))))))).. (-29.82 = -30.78 + 0.96)

| Location | 8,704,885 – 8,704,992 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -25.50 |
| Energy contribution | -26.91 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8704885 107 - 23771897 --GCUCGACU--CGAAUGUCAAGC---GAUGGCCAGCAACAUUGAAGCUCUUGGCCAAGAGUU---UGGCCCCAAACAAGUCAGCA--GAUGCGAUUGCUUACCAGCCAGAAUACA-GAA --(((.(((.--.....))).)))---..((((((((.........))...))))))....((---((((........((((.((.--...))))))........)))))).....-... ( -28.09) >DroSec_CAF1 10432 108 - 1 --GCUCGACU--CGAAUGUCAAGC---GAUGGCCAGCAGCAUUGAAGCUCGUGGCCAAGAGUU---UGGCCCCAAACAAGUCAGCA--GAUGCGAUUGCUUACCAGCCAGAAAAUAUAAA --(((.(((.--.....))).)))---..((((((..(((......)))..))))))....((---((((........((((.((.--...))))))........))))))......... ( -32.19) >DroSim_CAF1 11249 108 - 1 --GCUCGACU--CGAAUGUCAAGC---GAUGGCCAGCAGCAUUGAAGCUCUUGGCCAAGAGUU---UGGCCCCAAACAAGUCAGCA--GAUGCGAUUGCUUACCAGCCAGAAAAUAUGAA --(((.(((.--.....))).)))---..(((((((.(((......))).)))))))....((---((((........((((.((.--...))))))........))))))......... ( -33.79) >DroEre_CAF1 10415 105 - 1 --GCUCGACU--CGAAUGUCAAGC---GCUGGCCAGCAGCAUUGAAGCUCUUGGCCAAGAGUU---CGGCCCCAAACAAGUCAGCA--GAUGCGAUUGCUUAGCAGCCAGAAAAUAU--- --(((.(((.--.....))).)))---..(((((((.(((......))).)))))))....((---((((........((((.((.--...))))))........))).))).....--- ( -31.49) >DroYak_CAF1 10533 107 - 1 --GCUCGACUGUCGAAUGUCAAGC---GAUGGCCAGCAGCAUUGAAGCUCUUGGCCAAGAGUU---UGGCCCCAAACAAGUCAGCA--GAUGCGAUUGCUUAGCAGCCAGAAAAUAU--- --(((.((((((.....(((((((---..(((((((.(((......))).)))))))...)))---)))).....)).))))))).--..(((.........)))............--- ( -34.30) >DroAna_CAF1 10262 107 - 1 CAGCUCGACU--CGAAUGUCAAGCUUGGAUGGCCAAGAA-----------AUGGCCAAGAGUUUUCUGGCCAAACAGAAAUCAGCACAGAACGGUUUGCUUACCAGCCAGAAAAUCAGAA .((((.(((.--.....))).))))....((((((....-----------.)))))).((.(((((((((..........((......))..(((......))).))))))))))).... ( -33.10) >consensus __GCUCGACU__CGAAUGUCAAGC___GAUGGCCAGCAGCAUUGAAGCUCUUGGCCAAGAGUU___UGGCCCCAAACAAGUCAGCA__GAUGCGAUUGCUUACCAGCCAGAAAAUAUGAA ..(((((((........))).........(((((((.(((......))).))))))).))))....((((........((((.((......))))))........))))........... (-25.50 = -26.91 + 1.40)

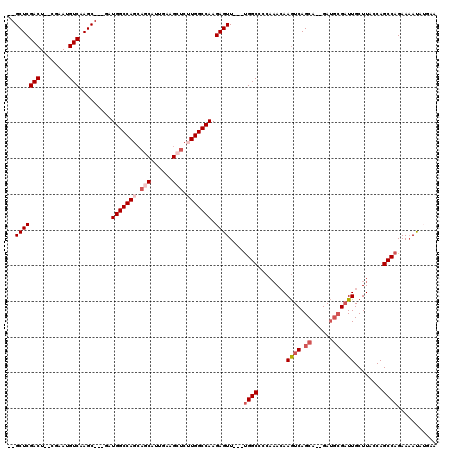
| Location | 8,704,922 – 8,705,021 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.68 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8704922 99 + 23771897 CUUGUUUGGGGCCAAACUCUUGGCCAAGAGCUUCAAUGUUGCUGGCCAUCGCUUGACAUUCG--AGUCGAGCGGUGAAUCACGGCAAGAAUUCUGGCAUCG .........((((((....))))))..(((((..(((.((((((..(((((((((((.....--.))))))))))).....))))))..)))..))).)). ( -39.10) >DroPse_CAF1 10667 88 + 1 CUGGUUUACGUCCGAGCUGUGGGCCA----------UAUUGCC-ACCUUCGGUUGACAUUCG--AGUCGCUUGGUGAAUCACCAAGAGAAUUCUUGACUUG ...(((.....(((((..((((....----------.....))-)).)))))..)))...((--((((.(((((((...)))))))((....)).)))))) ( -24.00) >DroSec_CAF1 10470 99 + 1 CUUGUUUGGGGCCAAACUCUUGGCCACGAGCUUCAAUGCUGCUGGCCAUCGCUUGACAUUCG--AGUCGAGCGGUGAAUCACGGCAAGAAUUCUGGCUUCG .......(((((((((.(((((((((..(((......)))..))))(((((((((((.....--.)))))))))))........))))).)).))))))). ( -42.30) >DroSim_CAF1 11287 99 + 1 CUUGUUUGGGGCCAAACUCUUGGCCAAGAGCUUCAAUGCUGCUGGCCAUCGCUUGACAUUCG--AGUCGAGCGGUGAAUCACGGCAAGAAUUCUGGCUUCG .......(((((((((.(((((((((..(((......)))..))))(((((((((((.....--.)))))))))))........))))).)).))))))). ( -42.70) >DroEre_CAF1 10450 99 + 1 CUUGUUUGGGGCCGAACUCUUGGCCAAGAGCUUCAAUGCUGCUGGCCAGCGCUUGACAUUCG--AGUCGAGCGGUGAAUCACGGCAAGAAUUCUGGCUUCG .......(((((((((.(((((((((..(((......)))..))))((.((((((((.....--.)))))))).))........))))).))).)))))). ( -42.60) >DroYak_CAF1 10568 101 + 1 CUUGUUUGGGGCCAAACUCUUGGCCAAGAGCUUCAAUGCUGCUGGCCAUCGCUUGACAUUCGACAGUCGAGCGGUGAAUCACGGCUAAAAUUCUGGCUUCG .......(((((((.....((((((.(((((......))).))...(((((((((((........)))))))))))......)))))).....))))))). ( -39.60) >consensus CUUGUUUGGGGCCAAACUCUUGGCCAAGAGCUUCAAUGCUGCUGGCCAUCGCUUGACAUUCG__AGUCGAGCGGUGAAUCACGGCAAGAAUUCUGGCUUCG .......(((((((......((((((..(((......)))..)))))).((((((((........))))))))....................))))))). (-30.70 = -30.68 + -0.02)

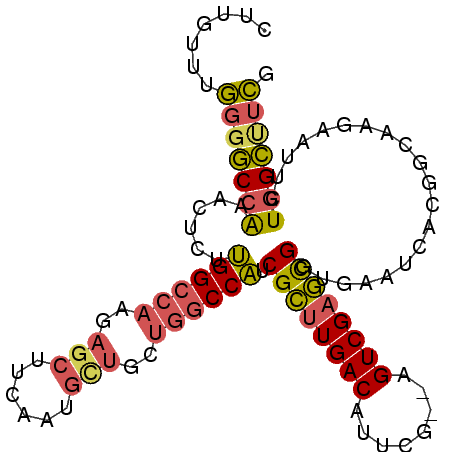
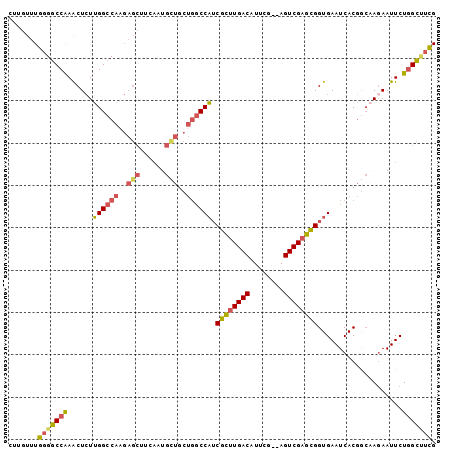
| Location | 8,704,922 – 8,705,021 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -25.03 |
| Energy contribution | -26.37 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 8704922 99 - 23771897 CGAUGCCAGAAUUCUUGCCGUGAUUCACCGCUCGACU--CGAAUGUCAAGCGAUGGCCAGCAACAUUGAAGCUCUUGGCCAAGAGUUUGGCCCCAAACAAG .(..((((((.(((((((((.((((((.((((.(((.--.....))).))))(((........)))))))..)).))).))))))))))))..)....... ( -29.50) >DroPse_CAF1 10667 88 - 1 CAAGUCAAGAAUUCUCUUGGUGAUUCACCAAGCGACU--CGAAUGUCAACCGAAGGU-GGCAAUA----------UGGCCCACAGCUCGGACGUAAACCAG ...............(((((((...))))))).(((.--.....)))..((((..((-(((....----------..).))))...))))........... ( -21.70) >DroSec_CAF1 10470 99 - 1 CGAAGCCAGAAUUCUUGCCGUGAUUCACCGCUCGACU--CGAAUGUCAAGCGAUGGCCAGCAGCAUUGAAGCUCGUGGCCAAGAGUUUGGCCCCAAACAAG ....((((((.(((((............((((.(((.--.....))).))))..(((((..(((......)))..)))))))))))))))).......... ( -33.30) >DroSim_CAF1 11287 99 - 1 CGAAGCCAGAAUUCUUGCCGUGAUUCACCGCUCGACU--CGAAUGUCAAGCGAUGGCCAGCAGCAUUGAAGCUCUUGGCCAAGAGUUUGGCCCCAAACAAG ....((((((.(((((............((((.(((.--.....))).))))..((((((.(((......))).))))))))))))))))).......... ( -34.90) >DroEre_CAF1 10450 99 - 1 CGAAGCCAGAAUUCUUGCCGUGAUUCACCGCUCGACU--CGAAUGUCAAGCGCUGGCCAGCAGCAUUGAAGCUCUUGGCCAAGAGUUCGGCCCCAAACAAG ....(((.((((((((............((((.(((.--.....))).))))..((((((.(((......))).))))))))))))))))).......... ( -35.90) >DroYak_CAF1 10568 101 - 1 CGAAGCCAGAAUUUUAGCCGUGAUUCACCGCUCGACUGUCGAAUGUCAAGCGAUGGCCAGCAGCAUUGAAGCUCUUGGCCAAGAGUUUGGCCCCAAACAAG ................((((.(((((..((((.(((........))).)))).(((((((.(((......))).))))))).))))))))).......... ( -33.30) >consensus CGAAGCCAGAAUUCUUGCCGUGAUUCACCGCUCGACU__CGAAUGUCAAGCGAUGGCCAGCAGCAUUGAAGCUCUUGGCCAAGAGUUUGGCCCCAAACAAG .(..(((.((((((((............((((.(((........))).))))..((((((.(((......))).)))))))))))))))))..)....... (-25.03 = -26.37 + 1.34)

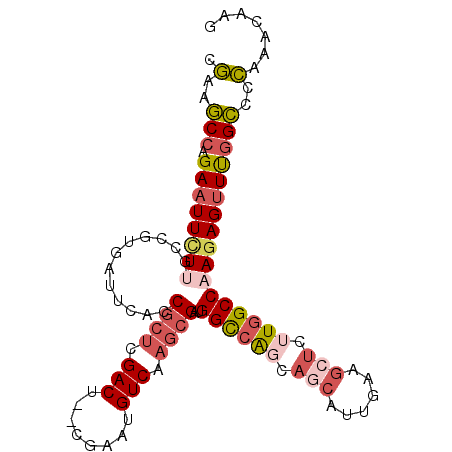
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:00 2006